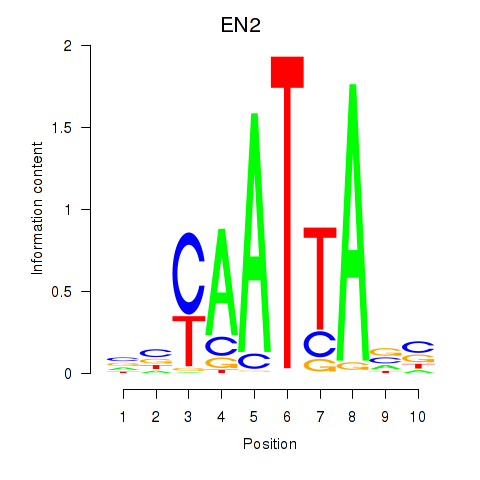
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for EN2_GBX2_LBX2
Z-value: 0.76
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN2
|
ENSG00000164778.4 | EN2 |
|
GBX2
|
ENSG00000168505.6 | GBX2 |
|
LBX2
|
ENSG00000179528.11 | LBX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GBX2 | hg19_v2_chr2_-_237076992_237077012 | -0.48 | 5.8e-02 | Click! |
| EN2 | hg19_v2_chr7_+_155250824_155250824 | -0.39 | 1.3e-01 | Click! |
Activity profile of EN2_GBX2_LBX2 motif
Sorted Z-values of EN2_GBX2_LBX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EN2_GBX2_LBX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_55562479 | 1.74 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 1.61 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr6_+_26199737 | 1.40 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr4_-_105416039 | 1.34 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr15_-_55562582 | 1.22 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr7_+_138145076 | 1.09 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr15_-_37393406 | 1.06 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr13_-_41593425 | 1.04 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr2_-_61697862 | 0.99 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr6_-_39693111 | 0.95 |
ENST00000373215.3 ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6 |
kinesin family member 6 |
| chr9_+_2159850 | 0.91 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_26199499 | 0.86 |
ENST00000377831.5 |
HIST1H3D |
histone cluster 1, H3d |
| chr6_+_135502501 | 0.85 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_-_32157947 | 0.84 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr16_-_29910853 | 0.75 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr6_-_111927062 | 0.73 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr3_+_167453493 | 0.62 |
ENST00000295777.5 ENST00000472747.2 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_-_242089677 | 0.61 |
ENST00000405260.1 |
PASK |
PAS domain containing serine/threonine kinase |
| chr11_+_67250490 | 0.60 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr12_+_28410128 | 0.60 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr18_+_59000815 | 0.59 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr18_-_31803435 | 0.58 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr3_-_141747950 | 0.57 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr18_+_55888767 | 0.56 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_-_33913708 | 0.56 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr13_-_36788718 | 0.53 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chrM_+_12331 | 0.53 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr1_-_190446759 | 0.52 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr21_-_43816052 | 0.50 |
ENST00000398405.1 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr17_-_39191107 | 0.50 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr6_-_26199471 | 0.48 |
ENST00000341023.1 |
HIST1H2AD |
histone cluster 1, H2ad |
| chr10_-_50970322 | 0.48 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr1_+_179923873 | 0.47 |
ENST00000367607.3 ENST00000491495.2 |
CEP350 |
centrosomal protein 350kDa |
| chr3_+_111718173 | 0.45 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr4_-_39979576 | 0.45 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr17_+_47448102 | 0.44 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr6_-_26250835 | 0.43 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr20_-_50722183 | 0.41 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr7_+_77428066 | 0.41 |
ENST00000422959.2 ENST00000307305.8 ENST00000424760.1 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr10_-_50970382 | 0.41 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr7_+_77428149 | 0.37 |
ENST00000415251.2 ENST00000275575.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr1_+_101003687 | 0.37 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr8_+_50824233 | 0.36 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr2_+_86668464 | 0.34 |
ENST00000409064.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr12_-_28123206 | 0.34 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr18_+_32173276 | 0.31 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr2_+_65216462 | 0.31 |
ENST00000234256.3 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr14_+_32798462 | 0.31 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr14_+_32798547 | 0.31 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr2_-_77749474 | 0.31 |
ENST00000409093.1 ENST00000409088.3 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
| chr12_-_28122980 | 0.29 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_14133053 | 0.28 |
ENST00000609686.1 |
GRIN2B |
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr4_-_116034979 | 0.28 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr1_+_171227069 | 0.28 |
ENST00000354841.4 |
FMO1 |
flavin containing monooxygenase 1 |
| chr7_+_100136811 | 0.28 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr14_-_51027838 | 0.28 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_156399184 | 0.27 |
ENST00000368243.1 ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr19_+_48949030 | 0.27 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr15_+_58430368 | 0.27 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr6_-_26032288 | 0.25 |
ENST00000244661.2 |
HIST1H3B |
histone cluster 1, H3b |
| chr2_+_196313239 | 0.25 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr3_+_115342349 | 0.25 |
ENST00000393780.3 |
GAP43 |
growth associated protein 43 |
| chr19_-_51522955 | 0.25 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr17_-_9929581 | 0.25 |
ENST00000437099.2 ENST00000396115.2 |
GAS7 |
growth arrest-specific 7 |
| chr18_-_31803169 | 0.25 |
ENST00000590712.1 |
NOL4 |
nucleolar protein 4 |
| chr12_+_52695617 | 0.24 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr1_+_62901968 | 0.23 |
ENST00000452143.1 ENST00000442679.1 ENST00000371146.1 |
USP1 |
ubiquitin specific peptidase 1 |
| chr1_-_152386732 | 0.23 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr11_+_75526212 | 0.23 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr6_-_76203345 | 0.22 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr1_+_110881945 | 0.22 |
ENST00000602849.1 ENST00000487146.2 |
RBM15 |
RNA binding motif protein 15 |
| chr12_-_52828147 | 0.22 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr6_+_34204642 | 0.21 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr10_-_48416849 | 0.21 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr1_+_214161272 | 0.21 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chrX_+_135730297 | 0.21 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr17_-_74163159 | 0.21 |
ENST00000591615.1 |
RNF157 |
ring finger protein 157 |
| chr19_-_59084647 | 0.21 |
ENST00000594234.1 ENST00000596039.1 |
MZF1 |
myeloid zinc finger 1 |
| chr2_+_171034646 | 0.20 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr2_+_47630108 | 0.20 |
ENST00000233146.2 ENST00000454849.1 ENST00000543555.1 |
MSH2 |
mutS homolog 2 |
| chr15_+_58430567 | 0.19 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr14_+_72052983 | 0.19 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr9_+_136501478 | 0.19 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chrX_-_53461305 | 0.19 |
ENST00000168216.6 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr4_-_26492076 | 0.18 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr4_-_120243545 | 0.18 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chrX_-_53461288 | 0.18 |
ENST00000375298.4 ENST00000375304.5 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr2_+_47630255 | 0.18 |
ENST00000406134.1 |
MSH2 |
mutS homolog 2 |
| chr7_-_25268104 | 0.18 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr19_+_4007644 | 0.18 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr17_-_48785216 | 0.17 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr8_+_105235572 | 0.17 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr12_+_7014064 | 0.17 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr7_+_127292234 | 0.16 |
ENST00000354725.3 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
| chr16_+_103816 | 0.16 |
ENST00000383018.3 ENST00000417493.1 |
SNRNP25 |
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr7_+_74072288 | 0.16 |
ENST00000443166.1 |
GTF2I |
general transcription factor IIi |
| chr9_-_116840728 | 0.16 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr19_-_19302931 | 0.15 |
ENST00000444486.3 ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B MEF2BNB MEF2B |
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr15_-_64673665 | 0.15 |
ENST00000300035.4 |
KIAA0101 |
KIAA0101 |
| chr20_-_18447667 | 0.15 |
ENST00000262547.5 ENST00000329494.5 ENST00000357236.4 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
| chr17_-_10017864 | 0.15 |
ENST00000323816.4 |
GAS7 |
growth arrest-specific 7 |
| chr8_-_42234745 | 0.14 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr15_-_64673630 | 0.13 |
ENST00000558008.1 ENST00000559519.1 ENST00000380258.2 |
KIAA0101 |
KIAA0101 |
| chr11_-_71823715 | 0.13 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr12_+_53662073 | 0.12 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr3_-_195538728 | 0.12 |
ENST00000349607.4 ENST00000346145.4 |
MUC4 |
mucin 4, cell surface associated |
| chr11_-_71823796 | 0.12 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr15_+_40733387 | 0.12 |
ENST00000416165.1 |
BAHD1 |
bromo adjacent homology domain containing 1 |
| chr9_+_80912059 | 0.12 |
ENST00000347159.2 ENST00000376588.3 |
PSAT1 |
phosphoserine aminotransferase 1 |
| chr12_-_53171128 | 0.12 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr4_-_174255536 | 0.12 |
ENST00000446922.2 |
HMGB2 |
high mobility group box 2 |
| chr19_+_11485333 | 0.11 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr4_-_89205879 | 0.10 |
ENST00000608933.1 ENST00000315194.4 ENST00000514204.1 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr3_+_152879985 | 0.10 |
ENST00000323534.2 |
RAP2B |
RAP2B, member of RAS oncogene family |
| chr12_-_14849470 | 0.10 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr14_-_104181771 | 0.10 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr9_+_12693336 | 0.10 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr3_-_52090461 | 0.09 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr12_+_53662110 | 0.09 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr3_+_173116225 | 0.09 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr4_-_174256276 | 0.09 |
ENST00000296503.5 |
HMGB2 |
high mobility group box 2 |
| chr3_-_195538760 | 0.09 |
ENST00000475231.1 |
MUC4 |
mucin 4, cell surface associated |
| chr1_+_153747746 | 0.09 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_+_111718036 | 0.08 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr2_-_163008903 | 0.08 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr1_+_62439037 | 0.08 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr19_+_51728316 | 0.08 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
| chr6_-_31088214 | 0.08 |
ENST00000376288.2 |
CDSN |
corneodesmosin |
| chr11_-_71823266 | 0.07 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr5_-_95297678 | 0.07 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr17_-_64225508 | 0.07 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr5_+_179159813 | 0.07 |
ENST00000292599.3 |
MAML1 |
mastermind-like 1 (Drosophila) |
| chr6_+_26020672 | 0.07 |
ENST00000357647.3 |
HIST1H3A |
histone cluster 1, H3a |
| chr19_-_14064114 | 0.07 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chr12_+_7013897 | 0.07 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr1_+_160160346 | 0.07 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_+_66300446 | 0.07 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_+_133292759 | 0.06 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chr8_-_25281747 | 0.05 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_+_53164833 | 0.04 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr7_-_73038867 | 0.04 |
ENST00000313375.3 ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL |
MLX interacting protein-like |
| chr8_+_24298597 | 0.04 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr15_-_31393910 | 0.04 |
ENST00000397795.2 ENST00000256552.6 ENST00000559179.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr12_+_7014126 | 0.04 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr4_-_89205705 | 0.04 |
ENST00000295908.7 ENST00000510548.2 ENST00000508256.1 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr15_+_89631647 | 0.04 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr6_+_131571535 | 0.03 |
ENST00000474850.2 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
| chr17_+_7792101 | 0.03 |
ENST00000358181.4 ENST00000330494.7 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr9_+_116225999 | 0.03 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_-_8093471 | 0.03 |
ENST00000389017.4 |
C17orf59 |
chromosome 17 open reading frame 59 |
| chr17_-_47045949 | 0.03 |
ENST00000357424.2 |
GIP |
gastric inhibitory polypeptide |
| chr3_-_119813264 | 0.03 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr8_+_24298531 | 0.02 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr16_+_69345243 | 0.02 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr5_+_145826867 | 0.02 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr6_-_161695074 | 0.02 |
ENST00000457520.2 ENST00000366906.5 ENST00000320285.4 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_-_23670752 | 0.01 |
ENST00000302271.6 ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr12_-_101604185 | 0.01 |
ENST00000536262.2 |
SLC5A8 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr6_-_31782813 | 0.01 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr3_+_130569429 | 0.01 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_-_73038822 | 0.01 |
ENST00000414749.2 ENST00000429400.2 ENST00000434326.1 |
MLXIPL |
MLX interacting protein-like |
| chr19_-_41870026 | 0.01 |
ENST00000243578.3 |
B9D2 |
B9 protein domain 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 4.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0032181 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.5 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 0.9 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 1.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.6 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.6 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.4 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 1.1 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |