Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for EOMES
Z-value: 1.02
Transcription factors associated with EOMES
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EOMES
|
ENSG00000163508.8 | EOMES |
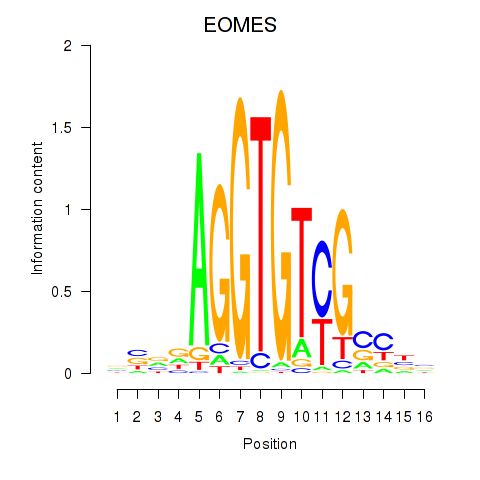
Activity profile of EOMES motif
Sorted Z-values of EOMES motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EOMES
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_28681950 | 1.65 |
ENST00000251081.6 |
DSC2 |
desmocollin 2 |
| chr1_-_207095324 | 1.48 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr1_-_207095212 | 1.39 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr16_+_202686 | 1.37 |
ENST00000252951.2 |
HBZ |
hemoglobin, zeta |
| chr1_-_184943610 | 1.22 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr3_+_50649302 | 1.06 |
ENST00000446044.1 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
| chrX_+_131157290 | 1.05 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_+_131157322 | 1.03 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr19_-_51456321 | 0.91 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr12_+_93965609 | 0.90 |
ENST00000549887.1 ENST00000551556.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr19_-_51456344 | 0.89 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr1_-_193029192 | 0.86 |
ENST00000417752.1 ENST00000367452.4 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
| chr11_-_107729887 | 0.84 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr17_-_39942940 | 0.81 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr2_-_99224915 | 0.78 |
ENST00000328709.3 ENST00000409997.1 |
COA5 |
cytochrome c oxidase assembly factor 5 |
| chr1_+_65775204 | 0.74 |
ENST00000371069.4 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr3_-_167452262 | 0.74 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr3_-_50649192 | 0.74 |
ENST00000443053.2 ENST00000348721.3 |
CISH |
cytokine inducible SH2-containing protein |
| chr1_-_149814478 | 0.74 |
ENST00000369161.3 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
| chr15_+_89182178 | 0.72 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr14_-_67878917 | 0.71 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr10_-_101190202 | 0.69 |
ENST00000543866.1 ENST00000370508.5 |
GOT1 |
glutamic-oxaloacetic transaminase 1, soluble |
| chr1_+_1981890 | 0.68 |
ENST00000378567.3 ENST00000468310.1 |
PRKCZ |
protein kinase C, zeta |
| chr4_-_144940477 | 0.66 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr22_-_37545972 | 0.66 |
ENST00000216223.5 |
IL2RB |
interleukin 2 receptor, beta |
| chr5_+_75699149 | 0.64 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr2_-_170430277 | 0.64 |
ENST00000438035.1 ENST00000453929.2 |
FASTKD1 |
FAST kinase domains 1 |
| chr1_+_150459873 | 0.64 |
ENST00000438568.2 ENST00000369054.2 ENST00000369064.3 ENST00000606933.1 |
TARS2 |
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr7_-_87505658 | 0.62 |
ENST00000341119.5 |
SLC25A40 |
solute carrier family 25, member 40 |
| chr2_-_235405168 | 0.61 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr15_+_89182156 | 0.60 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr7_-_99698338 | 0.60 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr15_+_41136586 | 0.60 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr2_-_163175133 | 0.59 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr7_+_87505544 | 0.58 |
ENST00000265728.1 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
| chr18_-_19284724 | 0.56 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr8_+_144099914 | 0.55 |
ENST00000521699.1 ENST00000520531.1 ENST00000520466.1 ENST00000521003.1 ENST00000522528.1 ENST00000522971.1 ENST00000519611.1 ENST00000521182.1 ENST00000519546.1 ENST00000523847.1 ENST00000522024.1 |
LY6E |
lymphocyte antigen 6 complex, locus E |
| chr1_+_70876891 | 0.54 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr1_+_70876926 | 0.49 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr15_+_89181974 | 0.49 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr8_+_144099896 | 0.49 |
ENST00000292494.6 ENST00000429120.2 |
LY6E |
lymphocyte antigen 6 complex, locus E |
| chr7_+_55177416 | 0.46 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr19_-_14629224 | 0.45 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr13_+_28194873 | 0.43 |
ENST00000302979.3 |
POLR1D |
polymerase (RNA) I polypeptide D, 16kDa |
| chr3_-_167452298 | 0.42 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr2_-_148778258 | 0.42 |
ENST00000392857.5 ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4 |
origin recognition complex, subunit 4 |
| chr13_+_28195988 | 0.42 |
ENST00000399697.3 ENST00000399696.1 |
POLR1D |
polymerase (RNA) I polypeptide D, 16kDa |
| chr8_+_86089619 | 0.42 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr1_-_205744205 | 0.42 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr20_-_18038521 | 0.41 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr6_+_31583761 | 0.41 |
ENST00000376049.4 |
AIF1 |
allograft inflammatory factor 1 |
| chr11_+_111896320 | 0.41 |
ENST00000531306.1 ENST00000537636.1 |
DLAT |
dihydrolipoamide S-acetyltransferase |
| chr1_+_213031570 | 0.40 |
ENST00000366971.4 |
FLVCR1 |
feline leukemia virus subgroup C cellular receptor 1 |
| chr4_-_151936865 | 0.39 |
ENST00000535741.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr7_-_37024665 | 0.39 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr11_-_57103327 | 0.39 |
ENST00000529002.1 ENST00000278412.2 |
SSRP1 |
structure specific recognition protein 1 |
| chr22_-_18257178 | 0.39 |
ENST00000342111.5 |
BID |
BH3 interacting domain death agonist |
| chr16_+_30064444 | 0.38 |
ENST00000395248.1 ENST00000566897.1 ENST00000568435.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr1_-_25747283 | 0.38 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr1_+_25598989 | 0.38 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chr12_+_6554021 | 0.38 |
ENST00000266557.3 |
CD27 |
CD27 molecule |
| chr16_-_67693846 | 0.37 |
ENST00000602850.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr1_+_19578033 | 0.37 |
ENST00000330263.4 |
MRTO4 |
mRNA turnover 4 homolog (S. cerevisiae) |
| chr6_-_111927062 | 0.37 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr3_+_141121164 | 0.37 |
ENST00000510338.1 ENST00000504673.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr2_-_148778323 | 0.37 |
ENST00000440042.1 ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4 |
origin recognition complex, subunit 4 |
| chr8_-_8318847 | 0.36 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr17_-_18585131 | 0.36 |
ENST00000443457.1 ENST00000583002.1 |
ZNF286B |
zinc finger protein 286B |
| chr9_+_136223414 | 0.36 |
ENST00000371964.4 |
SURF2 |
surfeit 2 |
| chrX_-_63005405 | 0.36 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr6_+_34204642 | 0.36 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr6_-_35888824 | 0.35 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr11_+_63706444 | 0.35 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr18_+_43753974 | 0.35 |
ENST00000282059.6 ENST00000321319.6 |
C18orf25 |
chromosome 18 open reading frame 25 |
| chr2_-_31637560 | 0.34 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr1_-_167906277 | 0.34 |
ENST00000271373.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr19_+_33865218 | 0.34 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr1_-_93426998 | 0.34 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr2_+_61108771 | 0.34 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr10_+_5454505 | 0.34 |
ENST00000355029.4 |
NET1 |
neuroepithelial cell transforming 1 |
| chr2_+_61108650 | 0.33 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr8_+_126010783 | 0.33 |
ENST00000521232.1 |
SQLE |
squalene epoxidase |
| chr1_-_205744574 | 0.33 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr19_-_39108568 | 0.33 |
ENST00000586296.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr3_-_182698381 | 0.33 |
ENST00000292782.4 |
DCUN1D1 |
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr17_-_62493131 | 0.33 |
ENST00000539111.2 |
POLG2 |
polymerase (DNA directed), gamma 2, accessory subunit |
| chr22_+_47070490 | 0.33 |
ENST00000408031.1 |
GRAMD4 |
GRAM domain containing 4 |
| chr1_-_245027833 | 0.33 |
ENST00000444376.2 |
HNRNPU |
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr17_+_79989937 | 0.33 |
ENST00000580965.1 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr19_+_49838653 | 0.33 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr3_+_111805182 | 0.32 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr9_+_37486005 | 0.32 |
ENST00000377792.3 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr11_+_111896090 | 0.32 |
ENST00000393051.1 |
DLAT |
dihydrolipoamide S-acetyltransferase |
| chr15_-_43622736 | 0.32 |
ENST00000544735.1 ENST00000567039.1 ENST00000305641.5 |
LCMT2 |
leucine carboxyl methyltransferase 2 |
| chr10_-_61900762 | 0.31 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_+_46018872 | 0.31 |
ENST00000583599.1 ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO |
pyridoxamine 5'-phosphate oxidase |
| chr11_+_73498898 | 0.31 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chr16_-_17564738 | 0.31 |
ENST00000261381.6 |
XYLT1 |
xylosyltransferase I |
| chr17_+_73521763 | 0.31 |
ENST00000167462.5 ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
| chr19_+_35645618 | 0.31 |
ENST00000392218.2 ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr17_+_9066252 | 0.31 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr1_-_213031418 | 0.30 |
ENST00000356684.3 ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1 |
FLVCR1 antisense RNA 1 (head to head) |
| chr1_+_233086326 | 0.30 |
ENST00000366628.5 ENST00000366627.4 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
| chr11_+_64808675 | 0.30 |
ENST00000529996.1 |
SAC3D1 |
SAC3 domain containing 1 |
| chrX_-_16730688 | 0.30 |
ENST00000359276.4 |
CTPS2 |
CTP synthase 2 |
| chr9_+_37485932 | 0.30 |
ENST00000377798.4 ENST00000442009.2 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr8_+_86089460 | 0.30 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr20_-_39317868 | 0.29 |
ENST00000373313.2 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr14_-_106963409 | 0.29 |
ENST00000390621.2 |
IGHV1-45 |
immunoglobulin heavy variable 1-45 |
| chr1_+_25598872 | 0.29 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr22_-_30662828 | 0.29 |
ENST00000403463.1 ENST00000215781.2 |
OSM |
oncostatin M |
| chr22_-_22221658 | 0.29 |
ENST00000544786.1 |
MAPK1 |
mitogen-activated protein kinase 1 |
| chr8_-_9760839 | 0.29 |
ENST00000519461.1 ENST00000517675.1 |
LINC00599 |
long intergenic non-protein coding RNA 599 |
| chr14_-_53162361 | 0.29 |
ENST00000395686.3 |
ERO1L |
ERO1-like (S. cerevisiae) |
| chr7_+_28452130 | 0.29 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr19_+_49496705 | 0.28 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr9_-_117853297 | 0.28 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr11_+_63655987 | 0.28 |
ENST00000509502.2 ENST00000512060.1 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr3_+_44379944 | 0.27 |
ENST00000396078.3 ENST00000342649.4 |
TCAIM |
T cell activation inhibitor, mitochondrial |
| chr20_-_57617831 | 0.27 |
ENST00000371033.5 ENST00000355937.4 |
SLMO2 |
slowmo homolog 2 (Drosophila) |
| chr5_-_112630598 | 0.27 |
ENST00000302475.4 |
MCC |
mutated in colorectal cancers |
| chr17_-_27405875 | 0.27 |
ENST00000359450.6 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_+_37940153 | 0.27 |
ENST00000373087.6 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
| chr12_-_99288536 | 0.27 |
ENST00000549797.1 ENST00000333732.7 ENST00000341752.7 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_-_94333784 | 0.27 |
ENST00000265986.6 |
IDE |
insulin-degrading enzyme |
| chr1_+_162531294 | 0.27 |
ENST00000367926.4 ENST00000271469.3 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr15_+_48623600 | 0.27 |
ENST00000558813.1 ENST00000331200.3 ENST00000558472.1 |
DUT |
deoxyuridine triphosphatase |
| chr19_-_39108552 | 0.27 |
ENST00000591517.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr19_+_34856141 | 0.27 |
ENST00000586425.1 |
GPI |
glucose-6-phosphate isomerase |
| chr19_+_35645817 | 0.27 |
ENST00000423817.3 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr4_-_26492076 | 0.26 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr1_+_150229554 | 0.26 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr11_+_117947724 | 0.26 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr4_+_17812525 | 0.26 |
ENST00000251496.2 |
NCAPG |
non-SMC condensin I complex, subunit G |
| chr1_-_167906020 | 0.26 |
ENST00000458574.1 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr16_+_30064411 | 0.26 |
ENST00000338110.5 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr22_-_22221900 | 0.26 |
ENST00000215832.6 ENST00000398822.3 |
MAPK1 |
mitogen-activated protein kinase 1 |
| chr1_-_245027766 | 0.26 |
ENST00000283179.9 |
HNRNPU |
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr16_-_67694129 | 0.25 |
ENST00000602320.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr11_+_64808368 | 0.25 |
ENST00000531072.1 ENST00000398846.1 |
SAC3D1 |
SAC3 domain containing 1 |
| chr17_-_17740287 | 0.25 |
ENST00000355815.4 ENST00000261646.5 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
| chr5_+_96271141 | 0.25 |
ENST00000231368.5 |
LNPEP |
leucyl/cystinyl aminopeptidase |
| chr8_+_73449625 | 0.25 |
ENST00000523207.1 |
KCNB2 |
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr2_+_45878790 | 0.24 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr9_+_139780942 | 0.24 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr3_-_167452614 | 0.24 |
ENST00000392750.2 ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10 |
programmed cell death 10 |
| chr5_-_33892204 | 0.24 |
ENST00000504830.1 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr16_-_87367879 | 0.24 |
ENST00000568879.1 |
RP11-178L8.4 |
RP11-178L8.4 |
| chr12_-_33049690 | 0.24 |
ENST00000070846.6 ENST00000340811.4 |
PKP2 |
plakophilin 2 |
| chr1_-_246729544 | 0.24 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr10_+_75910960 | 0.24 |
ENST00000539909.1 ENST00000286621.2 |
ADK |
adenosine kinase |
| chr18_+_2655692 | 0.24 |
ENST00000320876.6 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr1_+_160051319 | 0.24 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr11_+_117947782 | 0.23 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr3_+_132379154 | 0.23 |
ENST00000468022.1 ENST00000473651.1 ENST00000494238.2 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
| chr20_+_48807351 | 0.23 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr16_+_29817841 | 0.23 |
ENST00000322945.6 ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_-_75498308 | 0.23 |
ENST00000569540.1 |
TMEM170A |
transmembrane protein 170A |
| chr11_+_65479702 | 0.23 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr14_+_32546274 | 0.23 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr4_+_128554081 | 0.23 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr15_-_70388943 | 0.23 |
ENST00000559048.1 ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr12_-_63328817 | 0.23 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr10_-_58120996 | 0.23 |
ENST00000361148.6 ENST00000395405.1 ENST00000373944.3 |
ZWINT |
ZW10 interacting kinetochore protein |
| chr19_+_49496782 | 0.23 |
ENST00000601968.1 ENST00000596837.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr12_+_94071341 | 0.23 |
ENST00000542893.2 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_-_127842612 | 0.22 |
ENST00000417360.1 ENST00000322623.5 |
RUVBL1 |
RuvB-like AAA ATPase 1 |
| chr7_+_16793160 | 0.22 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr11_-_62439012 | 0.22 |
ENST00000532208.1 ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48 |
chromosome 11 open reading frame 48 |
| chr15_+_48623208 | 0.22 |
ENST00000559935.1 ENST00000559416.1 |
DUT |
deoxyuridine triphosphatase |
| chr21_-_46962379 | 0.22 |
ENST00000311124.4 ENST00000380010.4 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
| chr13_+_73302047 | 0.22 |
ENST00000377814.2 ENST00000377815.3 ENST00000390667.5 |
BORA |
bora, aurora kinase A activator |
| chrX_+_150151824 | 0.22 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr5_+_96211643 | 0.22 |
ENST00000437043.3 ENST00000510373.1 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
| chr15_-_90777277 | 0.22 |
ENST00000328649.6 |
CIB1 |
calcium and integrin binding 1 (calmyrin) |
| chr3_+_196466710 | 0.22 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr7_+_100026406 | 0.21 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chr17_+_4736627 | 0.21 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr3_-_167452703 | 0.21 |
ENST00000497056.2 ENST00000473645.2 |
PDCD10 |
programmed cell death 10 |
| chr1_-_21113105 | 0.21 |
ENST00000375000.1 ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3 |
heterochromatin protein 1, binding protein 3 |
| chr19_-_39108643 | 0.21 |
ENST00000396857.2 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr8_-_79717750 | 0.21 |
ENST00000263851.4 ENST00000379113.2 |
IL7 |
interleukin 7 |
| chr2_-_40006289 | 0.21 |
ENST00000260619.6 ENST00000454352.2 |
THUMPD2 |
THUMP domain containing 2 |
| chr1_+_209757051 | 0.21 |
ENST00000009105.1 ENST00000423146.1 ENST00000361322.2 |
CAMK1G |
calcium/calmodulin-dependent protein kinase IG |
| chr19_+_19303008 | 0.21 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr1_+_222791417 | 0.21 |
ENST00000344922.5 ENST00000344441.6 ENST00000344507.1 |
MIA3 |
melanoma inhibitory activity family, member 3 |
| chr1_+_215256467 | 0.20 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr6_+_41888926 | 0.20 |
ENST00000230340.4 |
BYSL |
bystin-like |
| chr7_+_29234028 | 0.20 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr16_-_75498553 | 0.20 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr15_+_44580899 | 0.20 |
ENST00000559222.1 ENST00000299957.6 |
CASC4 |
cancer susceptibility candidate 4 |
| chr18_-_47017956 | 0.20 |
ENST00000584895.1 ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32 RPL17 |
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr22_-_46373004 | 0.20 |
ENST00000339464.4 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
| chr10_+_85899196 | 0.19 |
ENST00000372134.3 |
GHITM |
growth hormone inducible transmembrane protein |
| chrX_-_71933888 | 0.19 |
ENST00000373542.4 ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
| chr3_+_44379611 | 0.19 |
ENST00000383746.3 ENST00000417237.1 |
TCAIM |
T cell activation inhibitor, mitochondrial |
| chr7_-_93520191 | 0.19 |
ENST00000545378.1 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr5_+_52083730 | 0.19 |
ENST00000282588.6 ENST00000274311.2 |
ITGA1 PELO |
integrin, alpha 1 pelota homolog (Drosophila) |
| chr19_+_50084561 | 0.19 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chrX_+_21958674 | 0.19 |
ENST00000404933.2 |
SMS |
spermine synthase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 1.8 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 2.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 1.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.7 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.2 | 0.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.6 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.2 | 0.6 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.5 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.2 | 0.6 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.2 | 0.5 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 1.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.6 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.6 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.3 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.2 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.1 | GO:0043132 | NAD transport(GO:0043132) |
| 0.1 | 0.9 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:1902202 | regulation of endothelial tube morphogenesis(GO:1901509) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.0 | GO:0035549 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 1.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0015866 | adenine transport(GO:0015853) ADP transport(GO:0015866) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) response to camptothecin(GO:1901563) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:0032898 | nerve growth factor processing(GO:0032455) neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.0 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.5 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 0.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.2 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.9 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 0.6 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.3 | 2.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 0.7 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.2 | 0.7 | GO:0070546 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 0.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 0.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.3 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 1.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 1.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |