Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for EP300
Z-value: 2.41
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | EP300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | -0.30 | 2.6e-01 | Click! |
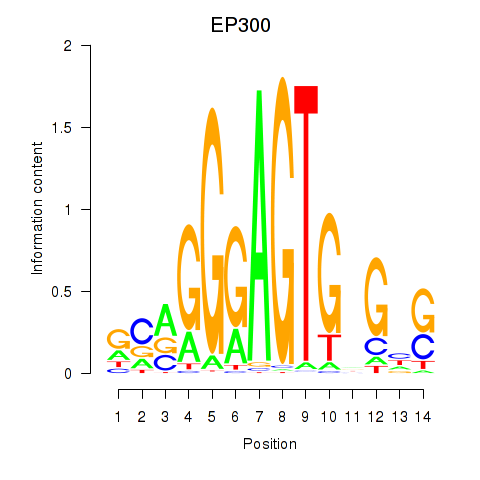
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_126849588 | 9.77 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chrX_-_140271249 | 8.94 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr12_+_6308881 | 8.82 |
ENST00000382518.1 ENST00000536586.1 |
CD9 |
CD9 molecule |
| chr12_+_6309517 | 7.69 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr18_-_21977748 | 7.24 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr14_-_69445968 | 5.75 |
ENST00000438964.2 |
ACTN1 |
actinin, alpha 1 |
| chr14_-_69446034 | 5.39 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr14_-_69445793 | 5.20 |
ENST00000538545.2 ENST00000394419.4 |
ACTN1 |
actinin, alpha 1 |
| chr1_-_20812690 | 4.92 |
ENST00000375078.3 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr5_-_39425222 | 4.86 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_36582857 | 4.82 |
ENST00000280527.2 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr5_-_39425290 | 4.73 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_-_110371720 | 4.43 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr3_-_149093499 | 4.20 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr8_+_30300119 | 4.13 |
ENST00000520191.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr1_+_10003486 | 4.06 |
ENST00000403197.1 ENST00000377205.1 |
NMNAT1 |
nicotinamide nucleotide adenylyltransferase 1 |
| chr14_+_105939276 | 3.91 |
ENST00000483017.3 |
CRIP2 |
cysteine-rich protein 2 |
| chr13_-_114567034 | 3.89 |
ENST00000327773.6 ENST00000357389.3 |
GAS6 |
growth arrest-specific 6 |
| chr3_-_32022733 | 3.71 |
ENST00000438237.2 ENST00000396556.2 |
OSBPL10 |
oxysterol binding protein-like 10 |
| chr13_-_40177261 | 3.67 |
ENST00000379589.3 |
LHFP |
lipoma HMGIC fusion partner |
| chr2_-_110371777 | 3.52 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr5_-_39425068 | 3.46 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_135229600 | 3.37 |
ENST00000370690.3 |
FHL1 |
four and a half LIM domains 1 |
| chr1_-_95007193 | 3.35 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chrX_+_134166333 | 3.30 |
ENST00000257013.7 |
FAM127A |
family with sequence similarity 127, member A |
| chr2_+_192110199 | 3.24 |
ENST00000304164.4 |
MYO1B |
myosin IB |
| chr2_+_192109911 | 3.22 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr11_+_131781290 | 3.18 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr1_+_20915409 | 3.18 |
ENST00000375071.3 |
CDA |
cytidine deaminase |
| chr16_+_66400533 | 3.14 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr2_-_110371412 | 3.11 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr8_+_15397732 | 3.09 |
ENST00000382020.4 ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3 |
tumor suppressor candidate 3 |
| chr9_+_19049372 | 3.08 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr6_+_29910301 | 3.08 |
ENST00000376809.5 ENST00000376802.2 |
HLA-A |
major histocompatibility complex, class I, A |
| chr20_-_56286479 | 3.00 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr8_-_134309335 | 3.00 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr10_-_16859361 | 2.97 |
ENST00000377921.3 |
RSU1 |
Ras suppressor protein 1 |
| chr11_-_46722117 | 2.79 |
ENST00000311956.4 |
ARHGAP1 |
Rho GTPase activating protein 1 |
| chr1_+_156030937 | 2.79 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr7_+_48128854 | 2.77 |
ENST00000436673.1 ENST00000429491.2 |
UPP1 |
uridine phosphorylase 1 |
| chrX_+_135229559 | 2.74 |
ENST00000394155.2 |
FHL1 |
four and a half LIM domains 1 |
| chr4_+_75311019 | 2.71 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr21_-_46293644 | 2.69 |
ENST00000330938.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr8_-_134309823 | 2.64 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr10_-_16859442 | 2.64 |
ENST00000602389.1 ENST00000345264.5 |
RSU1 |
Ras suppressor protein 1 |
| chr4_+_75480629 | 2.63 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr4_+_75310851 | 2.61 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr2_+_46524537 | 2.60 |
ENST00000263734.3 |
EPAS1 |
endothelial PAS domain protein 1 |
| chr15_-_73925651 | 2.59 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr19_+_16187085 | 2.55 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr1_-_94079648 | 2.45 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr2_-_208030647 | 2.43 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr21_-_46293586 | 2.39 |
ENST00000445724.2 ENST00000397887.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr12_-_8815404 | 2.37 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chrX_+_37208521 | 2.33 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr2_+_201171064 | 2.30 |
ENST00000451764.2 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr2_+_201171242 | 2.24 |
ENST00000360760.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr12_-_105629852 | 2.23 |
ENST00000551662.1 ENST00000553097.1 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr1_-_108742957 | 2.17 |
ENST00000565488.1 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_+_105086056 | 2.09 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr14_+_23340822 | 2.08 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr3_+_105085734 | 2.06 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr5_+_65892174 | 2.06 |
ENST00000404260.3 ENST00000403625.2 ENST00000406374.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_105630016 | 2.05 |
ENST00000258530.3 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr5_+_96079240 | 2.02 |
ENST00000515663.1 |
CAST |
calpastatin |
| chr17_-_79269067 | 1.94 |
ENST00000288439.5 ENST00000374759.3 |
SLC38A10 |
solute carrier family 38, member 10 |
| chr11_-_8986474 | 1.87 |
ENST00000525069.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr19_+_16296191 | 1.84 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr2_+_201171372 | 1.83 |
ENST00000409140.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr11_+_62623621 | 1.81 |
ENST00000535296.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr2_+_110371905 | 1.73 |
ENST00000356454.3 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
| chr3_-_79816965 | 1.71 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_45927097 | 1.68 |
ENST00000340192.7 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_-_244013384 | 1.68 |
ENST00000366539.1 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_150974494 | 1.66 |
ENST00000392811.2 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr22_-_43042968 | 1.62 |
ENST00000407623.3 ENST00000396303.3 ENST00000438270.1 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr2_+_30369807 | 1.58 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr11_+_62623512 | 1.55 |
ENST00000377892.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr2_-_110371664 | 1.53 |
ENST00000545389.1 ENST00000423520.1 |
SEPT10 |
septin 10 |
| chr12_-_110888103 | 1.52 |
ENST00000426440.1 ENST00000228825.7 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr4_-_186456766 | 1.51 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr11_+_62623544 | 1.49 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr4_-_186456652 | 1.47 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr6_-_110500905 | 1.46 |
ENST00000392587.2 |
WASF1 |
WAS protein family, member 1 |
| chr20_-_44485835 | 1.45 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr6_+_3000057 | 1.42 |
ENST00000397717.2 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr18_-_34408902 | 1.41 |
ENST00000593035.1 ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr16_+_66968343 | 1.41 |
ENST00000417689.1 ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2 |
carboxylesterase 2 |
| chr1_+_84543734 | 1.39 |
ENST00000370689.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_17434689 | 1.37 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr1_-_22263790 | 1.36 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr3_-_158450475 | 1.35 |
ENST00000237696.5 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr2_+_30369859 | 1.34 |
ENST00000402003.3 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr8_+_95732095 | 1.33 |
ENST00000414645.2 |
DPY19L4 |
dpy-19-like 4 (C. elegans) |
| chr9_+_82186872 | 1.30 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr11_+_2421718 | 1.26 |
ENST00000380996.5 ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4 |
tumor suppressing subtransferable candidate 4 |
| chr17_-_74099772 | 1.25 |
ENST00000411744.2 ENST00000332065.5 |
EXOC7 |
exocyst complex component 7 |
| chr18_-_61089665 | 1.24 |
ENST00000238497.5 |
VPS4B |
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr9_+_82186682 | 1.22 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr18_-_33709268 | 1.22 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr10_-_49813090 | 1.18 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr16_-_29465668 | 1.16 |
ENST00000569622.1 |
RP11-345J4.5 |
BolA-like protein 2 |
| chr22_-_29075853 | 1.16 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr1_-_43833628 | 1.15 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr11_-_104035088 | 1.13 |
ENST00000302251.5 |
PDGFD |
platelet derived growth factor D |
| chr3_+_130745769 | 1.10 |
ENST00000412440.2 |
NEK11 |
NIMA-related kinase 11 |
| chr1_+_43855560 | 1.09 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr6_-_122792919 | 1.09 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr3_-_133969437 | 1.08 |
ENST00000460933.1 ENST00000296084.4 |
RYK |
receptor-like tyrosine kinase |
| chrX_-_153141302 | 1.08 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chr11_-_8986279 | 1.07 |
ENST00000534025.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr6_-_10415218 | 1.07 |
ENST00000466073.1 ENST00000498450.1 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chrX_-_107018969 | 1.06 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr4_+_15704679 | 1.04 |
ENST00000382346.3 |
BST1 |
bone marrow stromal cell antigen 1 |
| chr17_-_74099795 | 1.04 |
ENST00000406660.3 ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7 |
exocyst complex component 7 |
| chr10_+_99400443 | 1.03 |
ENST00000370631.3 |
PI4K2A |
phosphatidylinositol 4-kinase type 2 alpha |
| chr19_+_11546093 | 1.02 |
ENST00000591462.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr12_+_54378923 | 1.02 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr12_+_53693466 | 1.02 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr17_-_7145475 | 1.02 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr3_+_107241783 | 1.01 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr18_+_3262415 | 0.99 |
ENST00000581193.1 ENST00000400175.5 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr7_-_5569588 | 0.98 |
ENST00000417101.1 |
ACTB |
actin, beta |
| chr11_-_8985927 | 0.98 |
ENST00000528117.1 ENST00000309134.5 |
TMEM9B |
TMEM9 domain family, member B |
| chr3_+_130745688 | 0.98 |
ENST00000510769.1 ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11 |
NIMA-related kinase 11 |
| chr8_-_103668114 | 0.97 |
ENST00000285407.6 |
KLF10 |
Kruppel-like factor 10 |
| chr10_+_114709999 | 0.97 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_11546153 | 0.97 |
ENST00000591946.1 ENST00000252455.2 ENST00000412601.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr10_-_111683183 | 0.97 |
ENST00000403138.2 ENST00000369683.1 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr17_+_37844331 | 0.97 |
ENST00000578199.1 ENST00000406381.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr5_+_119799927 | 0.96 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr19_-_1605424 | 0.95 |
ENST00000589880.1 ENST00000585671.1 ENST00000591899.3 |
UQCR11 |
ubiquinol-cytochrome c reductase, complex III subunit XI |
| chr5_+_174151536 | 0.94 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr20_+_43104508 | 0.93 |
ENST00000262605.4 ENST00000372904.3 |
TTPAL |
tocopherol (alpha) transfer protein-like |
| chr12_+_4699244 | 0.92 |
ENST00000540757.2 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr11_-_18610214 | 0.91 |
ENST00000300038.7 ENST00000396197.3 ENST00000320750.6 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chrX_-_100872911 | 0.91 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr2_-_3606206 | 0.91 |
ENST00000315212.3 |
RNASEH1 |
ribonuclease H1 |
| chr11_-_104972158 | 0.89 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr17_-_19266045 | 0.88 |
ENST00000395616.3 |
B9D1 |
B9 protein domain 1 |
| chr3_-_49761337 | 0.88 |
ENST00000535833.1 ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3 GMPPB |
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr12_+_52463751 | 0.88 |
ENST00000336854.4 ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44 |
chromosome 12 open reading frame 44 |
| chr6_+_2988847 | 0.86 |
ENST00000380472.3 ENST00000605901.1 ENST00000454015.1 |
NQO2 LINC01011 |
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr5_+_68530668 | 0.86 |
ENST00000506563.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chr19_+_11546440 | 0.84 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr10_-_69835099 | 0.83 |
ENST00000373700.4 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr1_+_40505891 | 0.81 |
ENST00000372797.3 ENST00000372802.1 ENST00000449311.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr16_+_68279207 | 0.81 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr12_+_53693812 | 0.80 |
ENST00000549488.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr2_+_231280908 | 0.80 |
ENST00000427101.2 ENST00000432979.1 |
SP100 |
SP100 nuclear antigen |
| chr10_-_49812997 | 0.78 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr16_+_68771128 | 0.76 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr3_+_171758344 | 0.76 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr2_+_231280954 | 0.76 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
| chr19_+_51226573 | 0.76 |
ENST00000250340.4 |
CLEC11A |
C-type lectin domain family 11, member A |
| chr5_+_68530697 | 0.75 |
ENST00000256443.3 ENST00000514676.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chrX_+_55478538 | 0.75 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chrX_+_95939711 | 0.75 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chrX_-_107019181 | 0.74 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chrX_+_54947229 | 0.74 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr22_+_20105259 | 0.73 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr17_-_19265855 | 0.73 |
ENST00000440841.1 ENST00000395615.1 ENST00000461069.2 |
B9D1 |
B9 protein domain 1 |
| chrX_+_23685563 | 0.72 |
ENST00000379341.4 |
PRDX4 |
peroxiredoxin 4 |
| chr7_-_32110451 | 0.72 |
ENST00000396191.1 ENST00000396182.2 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr12_+_116997186 | 0.71 |
ENST00000306985.4 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
| chrX_-_54522558 | 0.71 |
ENST00000375135.3 |
FGD1 |
FYVE, RhoGEF and PH domain containing 1 |
| chr11_-_104905840 | 0.69 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr19_-_56988677 | 0.69 |
ENST00000504904.3 ENST00000292069.6 |
ZNF667 |
zinc finger protein 667 |
| chrX_+_153665248 | 0.69 |
ENST00000447750.2 |
GDI1 |
GDP dissociation inhibitor 1 |
| chr16_+_31085714 | 0.67 |
ENST00000300850.5 ENST00000564189.1 ENST00000428260.1 |
ZNF646 |
zinc finger protein 646 |
| chr14_-_24711470 | 0.67 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr17_+_40118805 | 0.67 |
ENST00000591072.1 ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_-_180237445 | 0.66 |
ENST00000393340.3 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_+_21590549 | 0.65 |
ENST00000545178.1 ENST00000240651.9 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr12_-_57505121 | 0.63 |
ENST00000538913.2 ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr15_+_22892663 | 0.63 |
ENST00000313077.7 ENST00000561274.1 ENST00000560848.1 |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
| chr2_+_74153953 | 0.63 |
ENST00000264093.4 ENST00000348222.1 |
DGUOK |
deoxyguanosine kinase |
| chr10_-_99161033 | 0.62 |
ENST00000315563.6 ENST00000370992.4 ENST00000414986.1 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr18_+_3261883 | 0.61 |
ENST00000237500.5 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr17_+_4843679 | 0.61 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr4_+_56262115 | 0.60 |
ENST00000506198.1 ENST00000381334.5 ENST00000542052.1 |
TMEM165 |
transmembrane protein 165 |
| chr21_-_46707793 | 0.59 |
ENST00000331343.7 ENST00000349485.5 |
POFUT2 |
protein O-fucosyltransferase 2 |
| chrX_+_23685653 | 0.59 |
ENST00000379331.3 |
PRDX4 |
peroxiredoxin 4 |
| chrX_-_17879356 | 0.58 |
ENST00000331511.1 ENST00000415486.3 ENST00000545871.1 ENST00000451717.1 |
RAI2 |
retinoic acid induced 2 |
| chr22_+_29138013 | 0.58 |
ENST00000216027.3 ENST00000398941.2 |
HSCB |
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr17_-_26989136 | 0.58 |
ENST00000247020.4 |
SDF2 |
stromal cell-derived factor 2 |
| chr14_-_77923897 | 0.57 |
ENST00000343765.2 ENST00000327028.4 ENST00000556412.1 ENST00000557466.1 ENST00000448935.2 ENST00000553888.1 ENST00000557658.1 |
VIPAS39 |
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_-_161102421 | 0.57 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr15_+_74287009 | 0.57 |
ENST00000395135.3 |
PML |
promyelocytic leukemia |
| chr18_+_9334755 | 0.56 |
ENST00000262120.5 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
| chr16_-_30204987 | 0.56 |
ENST00000569282.1 ENST00000567436.1 |
BOLA2B |
bolA family member 2B |
| chr20_+_44035200 | 0.56 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr10_+_114710211 | 0.56 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_43855479 | 0.56 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr16_-_31085514 | 0.55 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr5_-_137878887 | 0.55 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr7_+_128470431 | 0.55 |
ENST00000325888.8 ENST00000346177.6 |
FLNC |
filamin C, gamma |
| chr15_+_23810853 | 0.55 |
ENST00000568252.1 |
MKRN3 |
makorin ring finger protein 3 |
| chr2_+_99225018 | 0.55 |
ENST00000357765.2 ENST00000409975.1 |
UNC50 |
unc-50 homolog (C. elegans) |
| chr6_+_3000218 | 0.54 |
ENST00000380441.1 ENST00000380455.4 ENST00000380454.4 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 22.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.7 | 13.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 2.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 3.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 4.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 17.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 4.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 4.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 7.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 4.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 2.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 2.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 13.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 16.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.8 | 9.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.7 | 4.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 16.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.6 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.4 | 1.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.4 | 4.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.7 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 2.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 1.6 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 1.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.3 | 3.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 1.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 4.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 4.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 4.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 4.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 8.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 5.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 7.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 5.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 4.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 11.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 4.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.4 | 4.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.8 | 3.3 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.8 | 4.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.7 | 2.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.7 | 16.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.6 | 2.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.6 | 13.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.6 | 4.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 4.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 1.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.5 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 2.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 1.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.3 | 3.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 1.4 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 6.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 2.2 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 4.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 1.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 2.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 7.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.9 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 7.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 1.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 4.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 3.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 14.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 4.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 11.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 2.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 4.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 5.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.4 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.0 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 3.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 8.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.5 | 16.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.3 | 3.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) renal albumin absorption(GO:0097018) oligodendrocyte apoptotic process(GO:0097252) regulation of renal albumin absorption(GO:2000532) |
| 1.1 | 3.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.1 | 9.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.1 | 3.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.0 | 16.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.0 | 4.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.9 | 2.8 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.7 | 2.8 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.7 | 8.9 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.7 | 5.3 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.7 | 2.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.6 | 1.7 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 | 1.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.5 | 5.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.5 | 4.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.5 | 4.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.4 | 1.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 1.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.4 | 1.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 1.6 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.4 | 1.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.4 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 3.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 1.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.3 | 2.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 1.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 1.0 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.3 | 2.5 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 0.9 | GO:0051795 | positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) activation of meiosis(GO:0090427) |
| 0.3 | 0.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 2.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 | 4.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.3 | 1.7 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.3 | 1.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 2.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 1.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 1.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 0.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 2.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 3.0 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 3.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.8 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 2.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 1.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 8.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 1.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 1.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 5.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 0.5 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.2 | 0.5 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 3.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 0.3 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.2 | 0.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 4.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 3.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 2.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 1.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 2.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.9 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.8 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 1.4 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 4.8 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 1.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) L-serine transport(GO:0015825) |
| 0.1 | 1.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.6 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 5.5 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.1 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.4 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 2.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 2.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 1.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 5.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 1.4 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 2.0 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 2.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.6 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 1.0 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 1.4 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.0 | 4.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 1.0 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 1.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.4 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 1.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.0 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.7 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 6.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 16.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 13.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 5.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 10.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 10.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |