Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
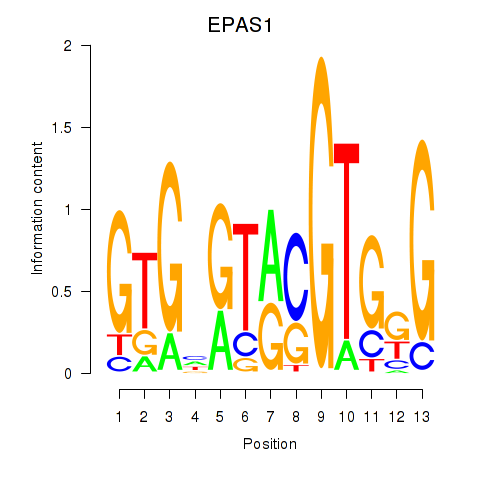
Results for EPAS1_BCL3
Z-value: 1.47
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.9 | EPAS1 |
|
BCL3
|
ENSG00000069399.8 | BCL3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EPAS1 | hg19_v2_chr2_+_46524537_46524553 | 0.85 | 2.8e-05 | Click! |
| BCL3 | hg19_v2_chr19_+_45251804_45251840 | 0.44 | 8.5e-02 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_7080227 | 6.00 |
ENST00000574330.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr3_-_32022733 | 4.44 |
ENST00000438237.2 ENST00000396556.2 |
OSBPL10 |
oxysterol binding protein-like 10 |
| chr5_+_52776228 | 4.43 |
ENST00000256759.3 |
FST |
follistatin |
| chr1_+_86046433 | 4.10 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr11_+_19798964 | 4.09 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr19_+_35629702 | 3.72 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr7_-_107642348 | 3.69 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr2_+_20646824 | 3.62 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr22_-_29075853 | 3.56 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr16_+_1578674 | 3.00 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr11_+_19799327 | 2.82 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr12_+_66217911 | 2.80 |
ENST00000403681.2 |
HMGA2 |
high mobility group AT-hook 2 |
| chr10_-_62761188 | 2.77 |
ENST00000357917.4 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr16_+_82068830 | 2.69 |
ENST00000199936.4 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr5_+_52776449 | 2.68 |
ENST00000396947.3 |
FST |
follistatin |
| chr9_+_103790991 | 2.57 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr17_-_7017559 | 2.53 |
ENST00000446679.2 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr2_-_238499303 | 2.38 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr5_+_52285144 | 2.38 |
ENST00000296585.5 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr16_+_72090053 | 2.36 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr20_+_34680620 | 2.28 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr8_-_134309335 | 2.21 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr3_-_99833333 | 2.17 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr6_+_31620191 | 2.17 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr16_-_70719925 | 2.12 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr3_-_134093275 | 2.08 |
ENST00000513145.1 ENST00000422605.2 |
AMOTL2 |
angiomotin like 2 |
| chr7_+_100770328 | 2.04 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr20_-_50385138 | 2.02 |
ENST00000338821.5 |
ATP9A |
ATPase, class II, type 9A |
| chr22_-_36236265 | 1.98 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr8_-_27462822 | 1.97 |
ENST00000522098.1 |
CLU |
clusterin |
| chr7_-_29234802 | 1.95 |
ENST00000449801.1 ENST00000409850.1 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr12_-_7261772 | 1.93 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr3_-_52479043 | 1.92 |
ENST00000231721.2 ENST00000475739.1 |
SEMA3G |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr4_+_74275057 | 1.88 |
ENST00000511370.1 |
ALB |
albumin |
| chr19_+_45449301 | 1.86 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr17_-_41623075 | 1.85 |
ENST00000545089.1 |
ETV4 |
ets variant 4 |
| chr8_-_18541603 | 1.82 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr19_+_45449266 | 1.69 |
ENST00000592257.1 |
APOC2 |
apolipoprotein C-II |
| chr4_+_38869410 | 1.69 |
ENST00000358869.2 |
FAM114A1 |
family with sequence similarity 114, member A1 |
| chr1_-_161193349 | 1.62 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr22_-_36236623 | 1.60 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr22_+_35776828 | 1.58 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr10_+_123923105 | 1.55 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr3_+_30647994 | 1.54 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr14_-_54420133 | 1.53 |
ENST00000559501.1 ENST00000558984.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr19_+_45449228 | 1.51 |
ENST00000252490.4 |
APOC2 |
apolipoprotein C-II |
| chr2_-_56150910 | 1.50 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr4_-_186732048 | 1.50 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_-_39425068 | 1.48 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_41623009 | 1.41 |
ENST00000393664.2 |
ETV4 |
ets variant 4 |
| chr17_-_41623691 | 1.35 |
ENST00000545954.1 |
ETV4 |
ets variant 4 |
| chr10_+_123923205 | 1.35 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_31620455 | 1.35 |
ENST00000437771.1 ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6 |
BCL2-associated athanogene 6 |
| chr6_-_31620403 | 1.30 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr20_-_36793774 | 1.27 |
ENST00000361475.2 |
TGM2 |
transglutaminase 2 |
| chr2_-_56150184 | 1.26 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr17_+_2699697 | 1.25 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chrX_-_99891796 | 1.23 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr3_+_30648066 | 1.23 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr5_-_42825983 | 1.21 |
ENST00000506577.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr2_-_238499337 | 1.20 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr2_-_220436248 | 1.16 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr17_-_41623259 | 1.15 |
ENST00000538265.1 ENST00000591713.1 |
ETV4 |
ets variant 4 |
| chr1_-_21671968 | 1.15 |
ENST00000415912.2 |
ECE1 |
endothelin converting enzyme 1 |
| chr11_-_70507901 | 1.14 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr19_+_45445491 | 1.13 |
ENST00000592954.1 ENST00000419266.2 ENST00000589057.1 |
APOC4 APOC4-APOC2 |
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr17_-_41623716 | 1.08 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr6_-_31620149 | 1.07 |
ENST00000435080.1 ENST00000375976.4 ENST00000441054.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr11_-_2170786 | 1.06 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr7_+_29519486 | 1.06 |
ENST00000409041.4 |
CHN2 |
chimerin 2 |
| chrX_+_102631248 | 1.06 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr19_+_35630022 | 1.06 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr17_+_72427477 | 1.06 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr22_-_36424458 | 1.04 |
ENST00000438146.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_+_203595903 | 1.03 |
ENST00000367218.3 ENST00000367219.3 ENST00000391954.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr17_+_1665253 | 1.02 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr10_+_111967345 | 1.01 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr2_-_238499131 | 1.00 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr3_-_134093395 | 1.00 |
ENST00000249883.5 |
AMOTL2 |
angiomotin like 2 |
| chr8_+_21915368 | 1.00 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr7_-_95064264 | 0.99 |
ENST00000536183.1 ENST00000433091.2 ENST00000222572.3 |
PON2 |
paraoxonase 2 |
| chr8_+_123793633 | 0.97 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr18_+_3449821 | 0.96 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chrX_+_105937068 | 0.96 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_+_35630628 | 0.95 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr2_-_188312971 | 0.94 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr19_-_43382142 | 0.94 |
ENST00000597058.1 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
| chr18_+_3449695 | 0.93 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_-_92413353 | 0.93 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr5_-_96143796 | 0.92 |
ENST00000296754.3 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr14_+_101193246 | 0.91 |
ENST00000331224.6 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr2_-_220435963 | 0.91 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr17_+_1665345 | 0.91 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr9_+_34652164 | 0.89 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr14_+_101193164 | 0.88 |
ENST00000341267.4 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr3_-_149688502 | 0.86 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr12_+_56473628 | 0.86 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr16_+_16326352 | 0.86 |
ENST00000399336.4 ENST00000263012.6 ENST00000538468.1 |
NOMO3 |
NODAL modulator 3 |
| chr19_-_36297348 | 0.84 |
ENST00000589835.1 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr19_+_41725088 | 0.84 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr11_-_89224508 | 0.83 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr2_+_201170770 | 0.83 |
ENST00000409988.3 ENST00000409385.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr1_-_240775447 | 0.83 |
ENST00000318160.4 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
| chr16_+_14927538 | 0.83 |
ENST00000287667.7 |
NOMO1 |
NODAL modulator 1 |
| chr10_+_123922941 | 0.83 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr5_-_16916624 | 0.82 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr4_-_102268484 | 0.81 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_+_123970052 | 0.81 |
ENST00000373823.3 |
GSN |
gelsolin |
| chr11_-_72433346 | 0.80 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr16_-_18573396 | 0.80 |
ENST00000543392.1 ENST00000381474.3 ENST00000330537.6 |
NOMO2 |
NODAL modulator 2 |
| chr3_+_37903432 | 0.77 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr1_+_203595689 | 0.76 |
ENST00000357681.5 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr18_+_29171689 | 0.76 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr2_+_102686820 | 0.75 |
ENST00000409929.1 ENST00000424272.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr15_+_96875657 | 0.75 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr19_+_54024251 | 0.75 |
ENST00000253144.9 |
ZNF331 |
zinc finger protein 331 |
| chr5_-_172756506 | 0.75 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr3_+_179370517 | 0.74 |
ENST00000263966.3 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chrX_-_108976521 | 0.74 |
ENST00000469796.2 ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
| chr16_+_68279207 | 0.74 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr8_+_21916710 | 0.73 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chr11_-_89224638 | 0.73 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr16_+_57662419 | 0.72 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr3_-_50340996 | 0.72 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr5_+_78365577 | 0.72 |
ENST00000518666.1 ENST00000521567.1 |
BHMT2 |
betaine--homocysteine S-methyltransferase 2 |
| chr2_+_85360499 | 0.71 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr12_+_56473939 | 0.71 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_78365536 | 0.70 |
ENST00000255192.3 |
BHMT2 |
betaine--homocysteine S-methyltransferase 2 |
| chr16_+_31483451 | 0.70 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr8_+_21916680 | 0.70 |
ENST00000358242.3 ENST00000415253.1 |
DMTN |
dematin actin binding protein |
| chr11_-_27722021 | 0.70 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chr3_-_49459865 | 0.69 |
ENST00000427987.1 |
AMT |
aminomethyltransferase |
| chr5_-_55290773 | 0.69 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr11_+_66278080 | 0.69 |
ENST00000318312.7 ENST00000526815.1 ENST00000537537.1 ENST00000525809.1 ENST00000455748.2 ENST00000393994.2 |
BBS1 |
Bardet-Biedl syndrome 1 |
| chr7_-_30029574 | 0.68 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr4_-_89152474 | 0.68 |
ENST00000515655.1 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr19_-_52227221 | 0.68 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr2_+_62900986 | 0.67 |
ENST00000405015.3 ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1 |
EH domain binding protein 1 |
| chr19_+_35532612 | 0.67 |
ENST00000600390.1 ENST00000597419.1 |
HPN |
hepsin |
| chr4_+_140586922 | 0.64 |
ENST00000265498.1 ENST00000506797.1 |
MGST2 |
microsomal glutathione S-transferase 2 |
| chr12_+_133758115 | 0.64 |
ENST00000541009.2 ENST00000592241.1 |
ZNF268 |
zinc finger protein 268 |
| chr15_-_60690932 | 0.63 |
ENST00000559818.1 |
ANXA2 |
annexin A2 |
| chrX_+_135229600 | 0.63 |
ENST00000370690.3 |
FHL1 |
four and a half LIM domains 1 |
| chr1_+_157963063 | 0.62 |
ENST00000360089.4 ENST00000368173.3 ENST00000392272.2 |
KIRREL |
kin of IRRE like (Drosophila) |
| chrX_+_107069063 | 0.61 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr3_+_52009110 | 0.61 |
ENST00000491470.1 |
ABHD14A |
abhydrolase domain containing 14A |
| chr11_-_89224488 | 0.61 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_-_49459878 | 0.60 |
ENST00000546031.1 ENST00000458307.2 ENST00000430521.1 |
AMT |
aminomethyltransferase |
| chr3_-_114035026 | 0.60 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr10_-_62704005 | 0.59 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr2_+_201171064 | 0.59 |
ENST00000451764.2 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr4_-_102268628 | 0.59 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_21978312 | 0.59 |
ENST00000359708.4 ENST00000290101.4 |
RAP1GAP |
RAP1 GTPase activating protein |
| chr1_+_17866290 | 0.58 |
ENST00000361221.3 ENST00000452522.1 ENST00000434513.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_+_29519662 | 0.58 |
ENST00000424025.2 ENST00000439711.2 ENST00000421775.2 |
CHN2 |
chimerin 2 |
| chr2_-_27341765 | 0.58 |
ENST00000405600.1 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
| chr5_-_146889619 | 0.57 |
ENST00000343218.5 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr2_+_201171372 | 0.57 |
ENST00000409140.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr19_+_11466062 | 0.56 |
ENST00000251473.5 ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
| chr19_+_45973120 | 0.56 |
ENST00000592811.1 ENST00000586615.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr20_+_30946106 | 0.56 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr19_+_11466167 | 0.56 |
ENST00000591608.1 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
| chr19_+_35630344 | 0.55 |
ENST00000455515.2 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr19_+_33685490 | 0.54 |
ENST00000253193.7 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
| chr10_+_115438920 | 0.54 |
ENST00000429617.1 ENST00000369331.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr22_+_24407642 | 0.53 |
ENST00000454754.1 ENST00000263119.5 |
CABIN1 |
calcineurin binding protein 1 |
| chr3_-_114866084 | 0.53 |
ENST00000357258.3 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_+_146402925 | 0.53 |
ENST00000302085.4 |
SMAD1 |
SMAD family member 1 |
| chr2_-_220083671 | 0.53 |
ENST00000439002.2 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr19_-_43709703 | 0.53 |
ENST00000599391.1 ENST00000244295.9 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr1_+_82266053 | 0.52 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr22_+_38142235 | 0.52 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chrX_+_135229559 | 0.52 |
ENST00000394155.2 |
FHL1 |
four and a half LIM domains 1 |
| chr11_+_114128522 | 0.52 |
ENST00000535401.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr21_-_43786634 | 0.52 |
ENST00000291527.2 |
TFF1 |
trefoil factor 1 |
| chr17_+_70117153 | 0.51 |
ENST00000245479.2 |
SOX9 |
SRY (sex determining region Y)-box 9 |
| chr14_+_93260569 | 0.51 |
ENST00000163416.2 |
GOLGA5 |
golgin A5 |
| chr1_+_214161272 | 0.51 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr2_-_220083692 | 0.51 |
ENST00000265316.3 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr6_+_36646435 | 0.51 |
ENST00000244741.5 ENST00000405375.1 ENST00000373711.2 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr12_+_49740700 | 0.50 |
ENST00000549441.2 ENST00000395069.3 |
DNAJC22 |
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr10_-_32217717 | 0.50 |
ENST00000396144.4 ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12 |
Rho GTPase activating protein 12 |
| chr17_-_78194147 | 0.50 |
ENST00000534910.1 ENST00000326317.6 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
| chr13_-_33780133 | 0.50 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr2_-_100759037 | 0.49 |
ENST00000317233.4 ENST00000423966.1 ENST00000416492.1 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr7_+_73442422 | 0.49 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chrX_+_37208521 | 0.48 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr12_+_57849048 | 0.48 |
ENST00000266646.2 |
INHBE |
inhibin, beta E |
| chr11_-_70507867 | 0.48 |
ENST00000412252.1 ENST00000409161.1 ENST00000409530.1 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr11_-_73472096 | 0.48 |
ENST00000541588.1 ENST00000336083.3 ENST00000540771.1 ENST00000310653.6 |
RAB6A |
RAB6A, member RAS oncogene family |
| chr22_+_24407776 | 0.48 |
ENST00000405822.2 |
CABIN1 |
calcineurin binding protein 1 |
| chr12_-_71533055 | 0.47 |
ENST00000552128.1 |
TSPAN8 |
tetraspanin 8 |
| chr15_+_66994561 | 0.47 |
ENST00000288840.5 |
SMAD6 |
SMAD family member 6 |
| chr14_-_105420241 | 0.47 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr2_-_27341872 | 0.47 |
ENST00000312734.4 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
| chr16_-_18468926 | 0.46 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr11_-_2950642 | 0.46 |
ENST00000314222.4 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
| chr22_+_24407847 | 0.46 |
ENST00000445422.1 ENST00000398319.2 |
CABIN1 |
calcineurin binding protein 1 |
| chr9_+_137533615 | 0.46 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr3_+_52009066 | 0.45 |
ENST00000458031.2 ENST00000463937.1 ENST00000273596.3 |
ACY1 ABHD14A-ACY1 ABHD14A |
aminoacylase 1 ABHD14A-ACY1 readthrough (NMD candidate) abhydrolase domain containing 14A |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.2 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.7 | 5.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 1.1 | 4.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 1.0 | 6.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.0 | 2.9 | GO:0031052 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.9 | 2.8 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.9 | 3.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.9 | 4.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.9 | 7.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.8 | 2.4 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.8 | 2.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.8 | 4.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.7 | 2.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.6 | 1.9 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 5.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.6 | 1.8 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.5 | 2.2 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.5 | 1.6 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.5 | 1.6 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.5 | 1.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.5 | 0.5 | GO:0072190 | ureter urothelium development(GO:0072190) ureter morphogenesis(GO:0072197) |
| 0.5 | 1.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.4 | 2.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.4 | 1.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.4 | 1.5 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 2.0 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 2.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 0.9 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.3 | 1.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.3 | 1.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 1.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 0.8 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 0.7 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 0.7 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 0.7 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.2 | 0.6 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 0.6 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 | 1.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 4.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.2 | 2.2 | GO:0090232 | peripheral nervous system myelin maintenance(GO:0032287) positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 | 4.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 0.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 2.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.7 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 2.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.2 | 0.8 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.2 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.9 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.1 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 8.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.4 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) |
| 0.1 | 1.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.4 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.3 | GO:0060214 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) endocardium formation(GO:0060214) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) gall bladder development(GO:0061010) |
| 0.1 | 0.5 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.1 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 2.9 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 1.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 1.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.8 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.3 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.3 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.2 | GO:0003306 | Wnt signaling pathway involved in heart development(GO:0003306) |
| 0.1 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.4 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 2.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.2 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.9 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0003183 | mitral valve morphogenesis(GO:0003183) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 3.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 2.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.2 | GO:0051462 | cortisol secretion(GO:0043400) positive regulation of corticotropin secretion(GO:0051461) regulation of cortisol secretion(GO:0051462) cellular response to cocaine(GO:0071314) positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.9 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 0.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.2 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 0.2 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.1 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.6 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 1.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.6 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.4 | GO:0090128 | regulation of synapse maturation(GO:0090128) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0045658 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 1.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.8 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 6.8 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.3 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 3.6 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.4 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0071205 | positive regulation of adenosine receptor signaling pathway(GO:0060168) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 1.2 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.1 | GO:0071621 | granulocyte chemotaxis(GO:0071621) |
| 0.0 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.5 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.1 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.3 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.5 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.0 | 5.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.0 | 2.9 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.7 | 2.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.7 | 2.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 1.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.6 | 1.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.5 | 2.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.5 | 2.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 1.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.5 | 1.4 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.4 | 1.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 1.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 7.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 2.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.3 | 1.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.3 | 5.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 2.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 0.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 4.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.6 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 2.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 6.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 6.3 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 4.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 3.1 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 6.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 3.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.0 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 3.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.8 | 5.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 2.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.6 | 6.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 2.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 2.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.5 | 4.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 2.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 3.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 6.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 1.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.3 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 2.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 2.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 2.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 2.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 6.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.0 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 5.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 8.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 3.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 2.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 7.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.8 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |