Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
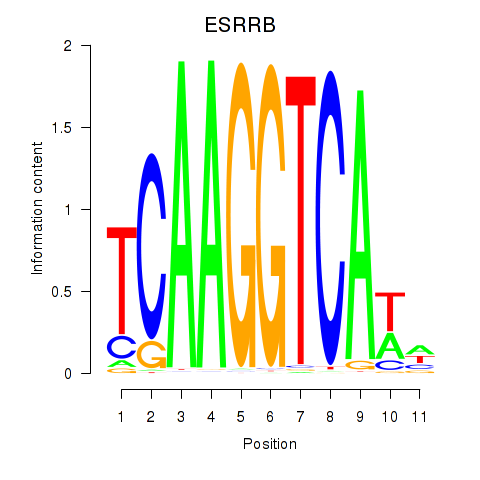
Results for ESRRB_ESRRG
Z-value: 1.54
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | ESRRB |
|
ESRRG
|
ENSG00000196482.12 | ESRRG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRG | hg19_v2_chr1_-_217250231_217250349 | -0.08 | 7.8e-01 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_235405168 | 5.60 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr21_+_42539701 | 4.62 |
ENST00000330333.6 ENST00000328735.6 ENST00000347667.5 |
BACE2 |
beta-site APP-cleaving enzyme 2 |
| chr1_+_46640750 | 3.29 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr17_-_39928106 | 3.17 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr2_-_106054952 | 3.16 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr2_-_161350305 | 3.03 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr17_-_27503770 | 2.88 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr3_-_123339418 | 2.88 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr8_+_86376081 | 2.84 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr3_-_123339343 | 2.81 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr15_+_43985725 | 2.67 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr17_-_39743139 | 2.63 |
ENST00000167586.6 |
KRT14 |
keratin 14 |
| chr4_-_143227088 | 2.54 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr15_+_43886057 | 2.44 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr9_-_35685452 | 2.33 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr22_+_29702996 | 2.32 |
ENST00000406549.3 ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr15_+_43985084 | 2.28 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr5_+_34757309 | 2.26 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr15_+_43885252 | 2.08 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr13_-_114567034 | 2.01 |
ENST00000327773.6 ENST00000357389.3 |
GAS6 |
growth arrest-specific 6 |
| chr22_+_30792846 | 1.93 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr22_+_30792980 | 1.86 |
ENST00000403484.1 ENST00000405717.3 ENST00000402592.3 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr2_+_192141611 | 1.69 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr11_+_12766583 | 1.65 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr16_-_47177874 | 1.61 |
ENST00000562435.1 |
NETO2 |
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr4_+_30721968 | 1.56 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr8_+_12808834 | 1.55 |
ENST00000400069.3 |
KIAA1456 |
KIAA1456 |
| chr9_-_130637244 | 1.54 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr8_+_12809093 | 1.52 |
ENST00000528753.2 |
KIAA1456 |
KIAA1456 |
| chr4_-_143767428 | 1.50 |
ENST00000513000.1 ENST00000509777.1 ENST00000503927.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_165600436 | 1.48 |
ENST00000367888.4 ENST00000367885.1 ENST00000367884.2 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr7_+_128399002 | 1.47 |
ENST00000493278.1 |
CALU |
calumenin |
| chr11_-_111794446 | 1.47 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr15_-_81616446 | 1.47 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr12_-_77272765 | 1.44 |
ENST00000547435.1 ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2 |
cysteine and glycine-rich protein 2 |
| chr7_+_55177416 | 1.40 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr7_-_151433342 | 1.27 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_+_48128816 | 1.27 |
ENST00000395564.4 |
UPP1 |
uridine phosphorylase 1 |
| chr7_-_151433393 | 1.27 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_-_23504176 | 1.24 |
ENST00000302291.4 |
LUZP1 |
leucine zipper protein 1 |
| chr11_-_47470591 | 1.23 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr11_-_47470703 | 1.21 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr7_+_48128854 | 1.19 |
ENST00000436673.1 ENST00000429491.2 |
UPP1 |
uridine phosphorylase 1 |
| chr12_-_21810726 | 1.15 |
ENST00000396076.1 |
LDHB |
lactate dehydrogenase B |
| chr11_-_47470682 | 1.14 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr17_+_900342 | 1.13 |
ENST00000327158.4 |
TIMM22 |
translocase of inner mitochondrial membrane 22 homolog (yeast) |
| chr2_-_220119280 | 1.11 |
ENST00000392088.2 |
TUBA4A |
tubulin, alpha 4a |
| chr3_-_167452262 | 1.11 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr12_-_21810765 | 1.11 |
ENST00000450584.1 ENST00000350669.1 |
LDHB |
lactate dehydrogenase B |
| chr7_+_48128194 | 1.11 |
ENST00000416681.1 ENST00000331803.4 ENST00000432131.1 |
UPP1 |
uridine phosphorylase 1 |
| chrX_-_13835147 | 1.10 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr7_+_48128316 | 1.09 |
ENST00000341253.4 |
UPP1 |
uridine phosphorylase 1 |
| chr10_+_71078595 | 1.09 |
ENST00000359426.6 |
HK1 |
hexokinase 1 |
| chr1_+_19923454 | 1.09 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr1_-_153600656 | 1.08 |
ENST00000339556.4 ENST00000440685.2 |
S100A13 |
S100 calcium binding protein A13 |
| chr12_+_54378923 | 1.00 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr9_+_139874683 | 1.00 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr11_-_6341724 | 0.99 |
ENST00000530979.1 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr1_+_152881014 | 0.97 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr7_+_79764104 | 0.97 |
ENST00000351004.3 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr16_+_28875126 | 0.96 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr12_-_53320245 | 0.96 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chrX_+_134166333 | 0.94 |
ENST00000257013.7 |
FAM127A |
family with sequence similarity 127, member A |
| chr8_-_18541603 | 0.94 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr3_-_38691119 | 0.94 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr2_+_220491973 | 0.93 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr12_-_28122980 | 0.89 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr2_-_238322770 | 0.85 |
ENST00000472056.1 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr1_-_144995002 | 0.85 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr2_-_238322800 | 0.84 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr11_+_66624527 | 0.84 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr2_+_170366203 | 0.82 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr12_+_119616447 | 0.80 |
ENST00000281938.2 |
HSPB8 |
heat shock 22kDa protein 8 |
| chr1_-_144995074 | 0.78 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr15_+_43809797 | 0.78 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr1_-_17380630 | 0.78 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_62623621 | 0.78 |
ENST00000535296.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr8_-_131028869 | 0.78 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr14_-_103987679 | 0.77 |
ENST00000553610.1 |
CKB |
creatine kinase, brain |
| chrX_+_49832231 | 0.77 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr4_-_71705082 | 0.75 |
ENST00000439371.1 ENST00000499044.2 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr11_+_57480046 | 0.75 |
ENST00000378312.4 ENST00000278422.4 |
TMX2 |
thioredoxin-related transmembrane protein 2 |
| chr10_+_75757863 | 0.74 |
ENST00000372755.3 ENST00000211998.4 ENST00000417648.2 |
VCL |
vinculin |
| chr1_-_145075847 | 0.74 |
ENST00000530740.1 ENST00000369359.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr9_-_14314518 | 0.71 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr10_+_17270214 | 0.71 |
ENST00000544301.1 |
VIM |
vimentin |
| chr11_+_62623512 | 0.68 |
ENST00000377892.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr6_-_97345689 | 0.67 |
ENST00000316149.7 |
NDUFAF4 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr1_-_201342364 | 0.67 |
ENST00000236918.7 ENST00000367317.4 ENST00000367315.2 ENST00000360372.4 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr11_+_62623544 | 0.67 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_+_236849754 | 0.66 |
ENST00000542672.1 ENST00000366578.4 |
ACTN2 |
actinin, alpha 2 |
| chr5_-_122372354 | 0.65 |
ENST00000306442.4 |
PPIC |
peptidylprolyl isomerase C (cyclophilin C) |
| chr11_+_67798114 | 0.65 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr8_-_110704014 | 0.64 |
ENST00000529190.1 ENST00000422135.1 ENST00000419099.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr1_-_207119738 | 0.64 |
ENST00000356495.4 |
PIGR |
polymeric immunoglobulin receptor |
| chr3_-_167452298 | 0.64 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr1_-_154164534 | 0.63 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr1_-_203144941 | 0.61 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr19_-_45826125 | 0.61 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr3_+_33318914 | 0.61 |
ENST00000484457.1 ENST00000538892.1 ENST00000538181.1 ENST00000446237.3 ENST00000507198.1 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
| chr1_+_202995611 | 0.60 |
ENST00000367240.2 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr2_+_217498105 | 0.60 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr17_-_73505961 | 0.59 |
ENST00000433559.2 |
CASKIN2 |
CASK interacting protein 2 |
| chr11_-_111957451 | 0.59 |
ENST00000504148.2 ENST00000541231.1 |
TIMM8B |
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr10_+_112257596 | 0.59 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr3_-_42845951 | 0.58 |
ENST00000418900.2 ENST00000430190.1 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
| chr8_-_145018080 | 0.58 |
ENST00000354589.3 |
PLEC |
plectin |
| chr20_+_1875942 | 0.58 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr3_-_10028366 | 0.58 |
ENST00000429759.1 |
EMC3 |
ER membrane protein complex subunit 3 |
| chr1_+_68150744 | 0.58 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr1_+_228270784 | 0.57 |
ENST00000541182.1 |
ARF1 |
ADP-ribosylation factor 1 |
| chr5_+_66254698 | 0.56 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr5_-_42811986 | 0.56 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr10_-_33625154 | 0.56 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr11_-_64014379 | 0.54 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr18_-_44336998 | 0.53 |
ENST00000315087.7 |
ST8SIA5 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr9_-_14313893 | 0.52 |
ENST00000380921.3 ENST00000380959.3 |
NFIB |
nuclear factor I/B |
| chr11_+_67798363 | 0.52 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_+_162531294 | 0.52 |
ENST00000367926.4 ENST00000271469.3 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr16_+_6069586 | 0.52 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_38219063 | 0.52 |
ENST00000584985.1 ENST00000264637.4 ENST00000450525.2 |
THRA |
thyroid hormone receptor, alpha |
| chr5_+_140227357 | 0.51 |
ENST00000378122.3 |
PCDHA9 |
protocadherin alpha 9 |
| chr5_+_80529104 | 0.50 |
ENST00000254035.4 ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr1_+_153388993 | 0.50 |
ENST00000368729.4 |
S100A7A |
S100 calcium binding protein A7A |
| chr1_-_33502528 | 0.50 |
ENST00000354858.6 |
AK2 |
adenylate kinase 2 |
| chr2_-_97304105 | 0.50 |
ENST00000599854.1 ENST00000441706.2 |
KANSL3 |
KAT8 regulatory NSL complex subunit 3 |
| chr9_-_100954910 | 0.50 |
ENST00000375077.4 |
CORO2A |
coronin, actin binding protein, 2A |
| chr20_-_48099182 | 0.50 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr17_+_48423453 | 0.49 |
ENST00000017003.2 ENST00000509778.1 ENST00000507602.1 |
XYLT2 |
xylosyltransferase II |
| chr16_+_6069072 | 0.49 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_-_57481278 | 0.49 |
ENST00000567751.1 ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1 |
cytokine induced apoptosis inhibitor 1 |
| chr4_+_56719782 | 0.49 |
ENST00000381295.2 ENST00000346134.7 ENST00000349598.6 |
EXOC1 |
exocyst complex component 1 |
| chr20_+_3776936 | 0.48 |
ENST00000439880.2 |
CDC25B |
cell division cycle 25B |
| chr4_-_170947522 | 0.48 |
ENST00000361618.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr9_-_14313641 | 0.47 |
ENST00000380953.1 |
NFIB |
nuclear factor I/B |
| chr1_-_1293904 | 0.47 |
ENST00000309212.6 ENST00000342753.4 ENST00000445648.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr4_-_74864386 | 0.47 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chr2_+_191792376 | 0.46 |
ENST00000409428.1 ENST00000409215.1 |
GLS |
glutaminase |
| chr9_-_14314066 | 0.46 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr11_-_33744487 | 0.46 |
ENST00000426650.2 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr19_+_45394477 | 0.46 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr15_-_72523924 | 0.46 |
ENST00000566809.1 ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM |
pyruvate kinase, muscle |
| chr4_+_41614909 | 0.46 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr8_-_131028660 | 0.45 |
ENST00000401979.2 ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_-_153433120 | 0.45 |
ENST00000368723.3 |
S100A7 |
S100 calcium binding protein A7 |
| chr15_-_41694640 | 0.45 |
ENST00000558719.1 ENST00000260361.4 ENST00000560978.1 |
NDUFAF1 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr7_+_44836276 | 0.45 |
ENST00000451562.1 ENST00000468812.1 ENST00000489459.1 ENST00000355968.6 |
PPIA |
peptidylprolyl isomerase A (cyclophilin A) |
| chr7_-_142232071 | 0.45 |
ENST00000390364.3 |
TRBV10-1 |
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr19_+_2249308 | 0.44 |
ENST00000592877.1 ENST00000221496.4 |
AMH |
anti-Mullerian hormone |
| chr7_-_98741714 | 0.44 |
ENST00000361125.1 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr1_-_211752073 | 0.44 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr8_+_136469684 | 0.43 |
ENST00000355849.5 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_-_145076186 | 0.43 |
ENST00000369348.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr8_-_131028641 | 0.43 |
ENST00000523509.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr2_-_97304009 | 0.43 |
ENST00000431828.1 ENST00000435669.1 ENST00000440133.1 ENST00000448075.1 |
KANSL3 |
KAT8 regulatory NSL complex subunit 3 |
| chr12_+_81101277 | 0.42 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr8_+_37887772 | 0.42 |
ENST00000338825.4 |
EIF4EBP1 |
eukaryotic translation initiation factor 4E binding protein 1 |
| chr16_-_4588469 | 0.42 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr15_-_72523454 | 0.41 |
ENST00000565154.1 ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM |
pyruvate kinase, muscle |
| chr1_-_20834586 | 0.41 |
ENST00000264198.3 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
| chr17_-_49198216 | 0.40 |
ENST00000262013.7 ENST00000357122.4 |
SPAG9 |
sperm associated antigen 9 |
| chr15_-_83621435 | 0.40 |
ENST00000450735.2 ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2 |
homer homolog 2 (Drosophila) |
| chr5_-_42812143 | 0.40 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr7_-_150777920 | 0.39 |
ENST00000353841.2 ENST00000297532.6 |
FASTK |
Fas-activated serine/threonine kinase |
| chr3_-_183735731 | 0.39 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_-_105029812 | 0.38 |
ENST00000482897.1 |
SRPK2 |
SRSF protein kinase 2 |
| chr21_-_27107344 | 0.38 |
ENST00000457143.2 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chrX_-_41449204 | 0.38 |
ENST00000378179.3 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr17_+_74372662 | 0.38 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr8_+_66556936 | 0.38 |
ENST00000262146.4 |
MTFR1 |
mitochondrial fission regulator 1 |
| chr5_+_149569520 | 0.38 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr12_-_57039739 | 0.37 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_129567282 | 0.37 |
ENST00000449886.1 ENST00000373464.4 ENST00000450858.1 |
ZBTB43 |
zinc finger and BTB domain containing 43 |
| chr19_-_55669093 | 0.37 |
ENST00000344887.5 |
TNNI3 |
troponin I type 3 (cardiac) |
| chr9_-_138853156 | 0.37 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr17_-_2169425 | 0.36 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr19_-_10697895 | 0.36 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr10_+_86088381 | 0.36 |
ENST00000224756.8 ENST00000372088.2 |
CCSER2 |
coiled-coil serine-rich protein 2 |
| chr11_-_64013663 | 0.36 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_+_32851487 | 0.35 |
ENST00000257836.3 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr21_-_27107283 | 0.35 |
ENST00000284971.3 ENST00000400099.1 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr20_-_44485835 | 0.35 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr5_+_32710736 | 0.34 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr17_+_46970134 | 0.34 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_-_111358372 | 0.34 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr16_-_2827128 | 0.34 |
ENST00000494946.2 ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2 |
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr10_+_5726764 | 0.33 |
ENST00000328090.5 ENST00000496681.1 |
FAM208B |
family with sequence similarity 208, member B |
| chr4_+_667686 | 0.33 |
ENST00000505477.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr7_-_150777949 | 0.32 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr3_-_50605077 | 0.32 |
ENST00000426034.1 ENST00000441239.1 |
C3orf18 |
chromosome 3 open reading frame 18 |
| chrX_+_47053208 | 0.32 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr2_+_220492116 | 0.31 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr1_+_201924619 | 0.31 |
ENST00000367287.4 |
TIMM17A |
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr16_-_19897455 | 0.31 |
ENST00000568214.1 ENST00000569479.1 |
GPRC5B |
G protein-coupled receptor, family C, group 5, member B |
| chr7_-_92463210 | 0.31 |
ENST00000265734.4 |
CDK6 |
cyclin-dependent kinase 6 |
| chr20_+_3776371 | 0.31 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr17_+_46970178 | 0.30 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr4_+_76439665 | 0.30 |
ENST00000508105.1 ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6 |
THAP domain containing 6 |
| chr4_-_71705027 | 0.30 |
ENST00000545193.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr7_-_98741642 | 0.30 |
ENST00000361368.2 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 1.2 | 4.7 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 1.0 | 2.9 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.9 | 2.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.7 | 5.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 2.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) renal albumin absorption(GO:0097018) oligodendrocyte apoptotic process(GO:0097252) regulation of renal albumin absorption(GO:2000532) |
| 0.6 | 3.2 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.6 | 3.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.6 | 6.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 4.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.5 | 2.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 1.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.4 | 2.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.7 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 3.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.0 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.3 | 2.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 1.7 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 0.7 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 0.6 | GO:0036090 | cleavage furrow ingression(GO:0036090) regulation of late endosome to lysosome transport(GO:1902822) |
| 0.2 | 0.5 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 0.7 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 0.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 2.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.9 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 2.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.2 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.1 | 0.5 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 1.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.6 | GO:1905040 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 1.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.4 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 1.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 4.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 1.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 1.5 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 2.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 1.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.7 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.8 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.9 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 1.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 4.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 3.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 1.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.8 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 3.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.5 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.4 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.6 | GO:1900449 | regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.9 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.5 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.4 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 1.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.4 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) |
| 0.0 | 0.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:0042325 | regulation of phosphorylation(GO:0042325) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 1.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.1 | 5.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.9 | 3.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.9 | 6.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 4.0 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 3.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.6 | 3.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 2.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 3.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 2.3 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.7 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.3 | 0.8 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 0.8 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.3 | 2.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 0.7 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 4.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 2.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 3.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 2.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.9 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 0.7 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 5.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.9 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 4.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 2.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.8 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 6.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 4.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 2.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 9.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 10.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 3.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 1.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 3.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 1.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 2.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 2.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 3.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 4.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 5.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.4 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 4.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.1 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 5.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 2.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 2.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |