Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
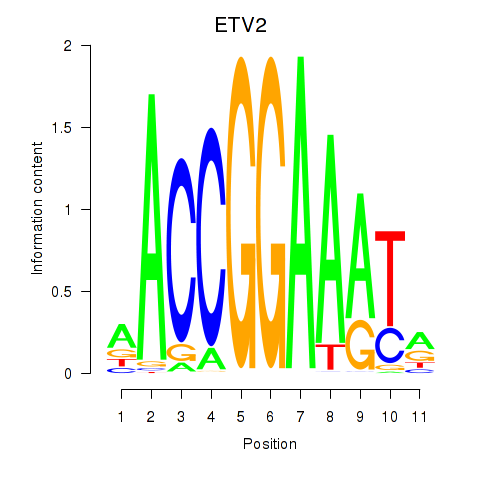
Results for ETV2
Z-value: 0.99
Transcription factors associated with ETV2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV2
|
ENSG00000105672.10 | ETV2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV2 | hg19_v2_chr19_+_36132631_36132695 | -0.51 | 4.6e-02 | Click! |
Activity profile of ETV2 motif
Sorted Z-values of ETV2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_238323007 | 1.88 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr2_-_238322800 | 1.87 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr2_-_238322770 | 1.78 |
ENST00000472056.1 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr5_+_125758813 | 1.35 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr11_-_119293872 | 1.33 |
ENST00000524970.1 |
THY1 |
Thy-1 cell surface antigen |
| chr5_+_125758865 | 1.32 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr17_+_1674982 | 1.32 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_+_22438009 | 1.26 |
ENST00000409417.1 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr8_-_119964434 | 1.23 |
ENST00000297350.4 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
| chr1_-_173174681 | 1.20 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr8_+_22437965 | 1.18 |
ENST00000409141.1 ENST00000265810.4 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr19_+_16296191 | 1.10 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr2_+_138721850 | 1.09 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr7_+_100770328 | 1.07 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_-_55523966 | 1.06 |
ENST00000474267.1 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr7_+_134528635 | 1.04 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr12_-_105629852 | 0.99 |
ENST00000551662.1 ENST00000553097.1 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr8_-_122653630 | 0.98 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr11_+_12399071 | 0.97 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr13_-_33760216 | 0.96 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr12_+_75874984 | 0.92 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr7_+_128379449 | 0.92 |
ENST00000479257.1 |
CALU |
calumenin |
| chr7_+_128379346 | 0.90 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr12_-_105630016 | 0.90 |
ENST00000258530.3 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_+_138722028 | 0.87 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr2_+_102721023 | 0.85 |
ENST00000409589.1 ENST00000409329.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chrX_+_51546103 | 0.77 |
ENST00000375772.3 |
MAGED1 |
melanoma antigen family D, 1 |
| chr7_-_19157248 | 0.76 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr4_-_57976544 | 0.76 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chrX_-_99891796 | 0.75 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr6_+_31620191 | 0.75 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr9_-_35689900 | 0.74 |
ENST00000378300.5 ENST00000329305.2 ENST00000360958.2 |
TPM2 |
tropomyosin 2 (beta) |
| chr12_+_75874580 | 0.69 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chrX_+_47444613 | 0.68 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr20_-_34542548 | 0.68 |
ENST00000305978.2 |
SCAND1 |
SCAN domain containing 1 |
| chr4_-_39529180 | 0.68 |
ENST00000515021.1 ENST00000510490.1 ENST00000316423.6 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr6_-_33239712 | 0.66 |
ENST00000436044.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_-_20987982 | 0.66 |
ENST00000375048.3 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr11_+_73882144 | 0.66 |
ENST00000328257.8 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr14_+_100842735 | 0.64 |
ENST00000554998.1 ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25 |
WD repeat domain 25 |
| chr11_+_73882311 | 0.64 |
ENST00000398427.4 ENST00000544401.1 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr7_-_107643674 | 0.64 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr1_+_99127265 | 0.62 |
ENST00000306121.3 |
SNX7 |
sorting nexin 7 |
| chr1_-_36789755 | 0.62 |
ENST00000270824.1 |
EVA1B |
eva-1 homolog B (C. elegans) |
| chr1_+_99127225 | 0.61 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr1_-_101360331 | 0.60 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr13_-_96705624 | 0.60 |
ENST00000376747.3 ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
| chrX_-_48931648 | 0.59 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr1_-_20987851 | 0.59 |
ENST00000464364.1 ENST00000602624.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr3_-_52002403 | 0.57 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr11_+_131781290 | 0.54 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr2_+_148602058 | 0.52 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr16_-_31085514 | 0.51 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr6_-_11382478 | 0.50 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chrX_+_55478538 | 0.50 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr1_-_154946825 | 0.49 |
ENST00000368453.4 ENST00000368450.1 ENST00000366442.2 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chrX_-_15872914 | 0.49 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr6_-_49430886 | 0.49 |
ENST00000274813.3 |
MUT |
methylmalonyl CoA mutase |
| chr1_-_43855444 | 0.49 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr2_-_231989808 | 0.47 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr15_+_90808919 | 0.47 |
ENST00000379095.3 |
NGRN |
neugrin, neurite outgrowth associated |
| chrX_+_54834791 | 0.47 |
ENST00000218439.4 ENST00000375058.1 ENST00000375060.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr14_+_45431379 | 0.47 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr12_-_76742183 | 0.47 |
ENST00000393262.3 |
BBS10 |
Bardet-Biedl syndrome 10 |
| chr8_+_38585704 | 0.47 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr5_+_147763539 | 0.46 |
ENST00000296701.6 ENST00000394370.3 |
FBXO38 |
F-box protein 38 |
| chr6_-_33239612 | 0.45 |
ENST00000482399.1 ENST00000445902.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr3_-_45883558 | 0.45 |
ENST00000445698.1 ENST00000296135.6 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr14_-_69864993 | 0.45 |
ENST00000555373.1 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
| chr1_-_20987889 | 0.45 |
ENST00000415136.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr11_+_70244510 | 0.44 |
ENST00000346329.3 ENST00000301843.8 ENST00000376561.3 |
CTTN |
cortactin |
| chr2_-_44588893 | 0.44 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr19_-_44860820 | 0.44 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr3_-_128879875 | 0.44 |
ENST00000418265.1 ENST00000393292.3 ENST00000273541.8 |
ISY1-RAB43 ISY1 |
ISY1-RAB43 readthrough ISY1 splicing factor homolog (S. cerevisiae) |
| chr11_-_46722117 | 0.44 |
ENST00000311956.4 |
ARHGAP1 |
Rho GTPase activating protein 1 |
| chr10_-_96122682 | 0.43 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr2_-_28113965 | 0.43 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chr5_+_147763498 | 0.43 |
ENST00000340253.5 |
FBXO38 |
F-box protein 38 |
| chrX_+_17755563 | 0.42 |
ENST00000380045.3 ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1 |
sex comb on midleg-like 1 (Drosophila) |
| chr19_-_50143452 | 0.42 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr2_-_44588624 | 0.42 |
ENST00000438314.1 ENST00000409936.1 |
PREPL |
prolyl endopeptidase-like |
| chr17_-_33905521 | 0.41 |
ENST00000225873.4 |
PEX12 |
peroxisomal biogenesis factor 12 |
| chr3_+_100428188 | 0.41 |
ENST00000418917.2 ENST00000490574.1 |
TFG |
TRK-fused gene |
| chr17_-_79818354 | 0.41 |
ENST00000576541.1 ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr17_-_37844267 | 0.41 |
ENST00000579146.1 ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3 |
post-GPI attachment to proteins 3 |
| chr1_-_43855479 | 0.41 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr17_+_37844331 | 0.41 |
ENST00000578199.1 ENST00000406381.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr1_-_155990580 | 0.41 |
ENST00000531917.1 ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2 |
signal sequence receptor, beta (translocon-associated protein beta) |
| chr6_-_83902933 | 0.40 |
ENST00000512866.1 ENST00000510258.1 ENST00000503094.1 ENST00000283977.4 ENST00000513973.1 ENST00000508748.1 |
PGM3 |
phosphoglucomutase 3 |
| chr15_-_65809581 | 0.40 |
ENST00000341861.5 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr6_-_30710447 | 0.40 |
ENST00000456573.2 |
FLOT1 |
flotillin 1 |
| chrX_+_70586140 | 0.40 |
ENST00000276072.3 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr3_-_28390581 | 0.40 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr14_-_75179774 | 0.39 |
ENST00000555249.1 ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1 AC007956.1 |
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr3_-_185542761 | 0.39 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_55897297 | 0.39 |
ENST00000431533.2 ENST00000428193.2 ENST00000558815.1 ENST00000560583.1 ENST00000560055.1 ENST00000559463.1 |
RPL28 |
ribosomal protein L28 |
| chr2_-_44588679 | 0.39 |
ENST00000409411.1 |
PREPL |
prolyl endopeptidase-like |
| chr17_+_7482785 | 0.38 |
ENST00000250092.6 ENST00000380498.6 ENST00000584502.1 |
CD68 |
CD68 molecule |
| chr3_+_69985734 | 0.38 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_43855560 | 0.38 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr19_-_4400415 | 0.38 |
ENST00000598564.1 ENST00000417295.2 ENST00000269886.3 |
SH3GL1 |
SH3-domain GRB2-like 1 |
| chr2_+_54198210 | 0.37 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr16_+_30210552 | 0.37 |
ENST00000338971.5 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr6_-_30710510 | 0.37 |
ENST00000376389.3 |
FLOT1 |
flotillin 1 |
| chr19_+_19496624 | 0.36 |
ENST00000494516.2 ENST00000360315.3 ENST00000252577.5 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr2_-_44588694 | 0.36 |
ENST00000409957.1 |
PREPL |
prolyl endopeptidase-like |
| chr20_+_16710606 | 0.36 |
ENST00000377943.5 ENST00000246071.6 |
SNRPB2 |
small nuclear ribonucleoprotein polypeptide B |
| chr15_-_74284613 | 0.36 |
ENST00000316911.6 ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1 |
stomatin (EPB72)-like 1 |
| chr3_-_185542817 | 0.36 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr20_+_19867150 | 0.36 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr4_-_39529049 | 0.35 |
ENST00000501493.2 ENST00000509391.1 ENST00000507089.1 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr20_-_20033052 | 0.35 |
ENST00000536226.1 |
CRNKL1 |
crooked neck pre-mRNA splicing factor 1 |
| chr11_-_6502580 | 0.35 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr4_+_2813946 | 0.34 |
ENST00000442312.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr5_-_137090028 | 0.34 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr20_+_44044717 | 0.33 |
ENST00000279036.6 ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT |
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr17_+_42264395 | 0.33 |
ENST00000587989.1 ENST00000590235.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr3_-_49055991 | 0.33 |
ENST00000441576.2 ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3 |
DALR anticodon binding domain containing 3 |
| chr18_-_812517 | 0.32 |
ENST00000584307.1 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr17_+_42264322 | 0.32 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr5_+_82373317 | 0.31 |
ENST00000282268.3 ENST00000338635.6 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr17_-_56429500 | 0.31 |
ENST00000225504.3 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
| chr20_+_3190006 | 0.30 |
ENST00000380113.3 ENST00000455664.2 ENST00000399838.3 |
ITPA |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr19_+_52693259 | 0.30 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr10_-_15902449 | 0.30 |
ENST00000277632.3 |
FAM188A |
family with sequence similarity 188, member A |
| chr2_+_231729615 | 0.30 |
ENST00000326427.6 ENST00000335005.6 ENST00000326407.6 |
ITM2C |
integral membrane protein 2C |
| chr11_+_43380459 | 0.29 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr6_+_83903061 | 0.29 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr1_+_84609944 | 0.29 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_+_3493611 | 0.29 |
ENST00000407558.4 ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr11_-_6502534 | 0.29 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr20_-_54967187 | 0.29 |
ENST00000422322.1 ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA |
aurora kinase A |
| chr4_+_74275057 | 0.29 |
ENST00000511370.1 |
ALB |
albumin |
| chr8_+_74903580 | 0.29 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr6_-_31620403 | 0.29 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr5_+_82373379 | 0.29 |
ENST00000396027.4 ENST00000511817.1 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr12_+_6833237 | 0.28 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr16_+_28722684 | 0.28 |
ENST00000331666.6 ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C |
eukaryotic translation initiation factor 3, subunit C |
| chr16_+_23569021 | 0.28 |
ENST00000567212.1 ENST00000567264.1 |
UBFD1 |
ubiquitin family domain containing 1 |
| chr3_-_184429735 | 0.28 |
ENST00000317897.3 |
MAGEF1 |
melanoma antigen family F, 1 |
| chr5_-_130970723 | 0.28 |
ENST00000308008.6 ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr12_+_108908962 | 0.28 |
ENST00000552695.1 ENST00000552758.1 ENST00000361549.2 |
FICD |
FIC domain containing |
| chr12_-_47473642 | 0.28 |
ENST00000266581.4 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr17_-_53809473 | 0.27 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr16_+_29471210 | 0.27 |
ENST00000360423.7 |
SULT1A4 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr21_-_36421535 | 0.27 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr7_-_27169801 | 0.27 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr17_+_79935464 | 0.27 |
ENST00000581647.1 ENST00000580534.1 ENST00000579684.1 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr12_-_47473557 | 0.27 |
ENST00000321382.3 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr8_-_42397037 | 0.27 |
ENST00000342228.3 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
| chr18_+_22006580 | 0.27 |
ENST00000284202.4 |
IMPACT |
impact RWD domain protein |
| chr8_-_101734308 | 0.27 |
ENST00000519004.1 ENST00000519363.1 ENST00000520142.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr4_-_103749205 | 0.27 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_56065484 | 0.27 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr19_-_40596767 | 0.27 |
ENST00000599972.1 ENST00000450241.2 ENST00000595687.2 |
ZNF780A |
zinc finger protein 780A |
| chr16_+_28722809 | 0.27 |
ENST00000566866.1 |
EIF3C |
eukaryotic translation initiation factor 3, subunit C |
| chr14_+_75348592 | 0.26 |
ENST00000334220.4 |
DLST |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr1_+_229440129 | 0.26 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr15_-_90234006 | 0.26 |
ENST00000300056.3 ENST00000559170.1 |
PEX11A |
peroxisomal biogenesis factor 11 alpha |
| chr6_+_142622991 | 0.26 |
ENST00000230173.6 ENST00000367608.2 |
GPR126 |
G protein-coupled receptor 126 |
| chr21_-_36421626 | 0.26 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr20_-_48532046 | 0.26 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr2_+_128848740 | 0.26 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr16_-_28415162 | 0.26 |
ENST00000398944.3 ENST00000398943.3 ENST00000380876.4 |
EIF3CL |
eukaryotic translation initiation factor 3, subunit C-like |
| chr1_+_17906970 | 0.26 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr2_+_128848881 | 0.25 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr7_-_76829125 | 0.25 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr19_+_52901094 | 0.25 |
ENST00000391788.2 ENST00000436397.1 ENST00000391787.2 ENST00000360465.3 ENST00000494167.2 ENST00000493272.1 |
ZNF528 |
zinc finger protein 528 |
| chr6_+_30525051 | 0.25 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chrX_+_54947229 | 0.25 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr15_-_65810042 | 0.25 |
ENST00000321147.6 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr1_-_165738085 | 0.25 |
ENST00000464650.1 ENST00000392129.6 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr1_-_28969517 | 0.25 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr16_+_82090028 | 0.25 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr4_-_103749179 | 0.25 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr20_-_5093503 | 0.25 |
ENST00000379277.2 |
TMEM230 |
transmembrane protein 230 |
| chr7_-_6746474 | 0.25 |
ENST00000394917.3 ENST00000405858.1 ENST00000342651.5 |
ZNF12 |
zinc finger protein 12 |
| chr20_+_33104199 | 0.25 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr4_-_103749105 | 0.25 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr15_-_65809991 | 0.24 |
ENST00000559526.1 ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr3_+_100428316 | 0.24 |
ENST00000479672.1 ENST00000476228.1 ENST00000463568.1 |
TFG |
TRK-fused gene |
| chrX_-_153775426 | 0.24 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr10_+_5726764 | 0.24 |
ENST00000328090.5 ENST00000496681.1 |
FAM208B |
family with sequence similarity 208, member B |
| chr2_-_136743169 | 0.24 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr12_+_6833437 | 0.24 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr20_-_49575081 | 0.24 |
ENST00000371588.5 ENST00000371582.4 |
DPM1 |
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr6_+_30524663 | 0.24 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr12_-_110888103 | 0.23 |
ENST00000426440.1 ENST00000228825.7 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr3_+_57541975 | 0.23 |
ENST00000487257.1 ENST00000311180.8 |
PDE12 |
phosphodiesterase 12 |
| chr1_+_40505891 | 0.23 |
ENST00000372797.3 ENST00000372802.1 ENST00000449311.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr16_+_57702099 | 0.23 |
ENST00000333493.4 ENST00000327655.6 |
GPR97 |
G protein-coupled receptor 97 |
| chr20_-_45142154 | 0.23 |
ENST00000347606.4 ENST00000457685.2 |
ZNF334 |
zinc finger protein 334 |
| chr8_-_101734170 | 0.23 |
ENST00000522387.1 ENST00000518196.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr2_+_162016827 | 0.23 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_+_120770645 | 0.22 |
ENST00000443902.2 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr12_-_47473425 | 0.22 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr18_-_812231 | 0.22 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr20_-_48532019 | 0.22 |
ENST00000289431.5 |
SPATA2 |
spermatogenesis associated 2 |
| chr17_+_7487146 | 0.22 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr11_-_47270341 | 0.22 |
ENST00000529444.1 ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2 |
acid phosphatase 2, lysosomal |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 4.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.3 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 1.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.6 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 7.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0043028 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0000406 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 0.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 0.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.7 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 2.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 2.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) lateral loop(GO:0043219) |
| 0.0 | 2.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 1.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 1.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.4 | 1.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.4 | 1.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.1 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.3 | 2.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 1.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.3 | 1.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 0.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 0.8 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.2 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.0 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 1.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.7 | GO:0034445 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 0.6 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 1.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.2 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.8 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.3 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.5 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.5 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.3 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.5 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.1 | 0.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.3 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:2000395 | ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.0 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.3 | GO:0071494 | positive regulation of translational initiation in response to stress(GO:0032058) cellular response to UV-C(GO:0071494) |
| 0.1 | 5.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.2 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 2.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0030397 | mitotic nuclear envelope disassembly(GO:0007077) membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.0 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.4 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.0 | 0.5 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.8 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |