Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
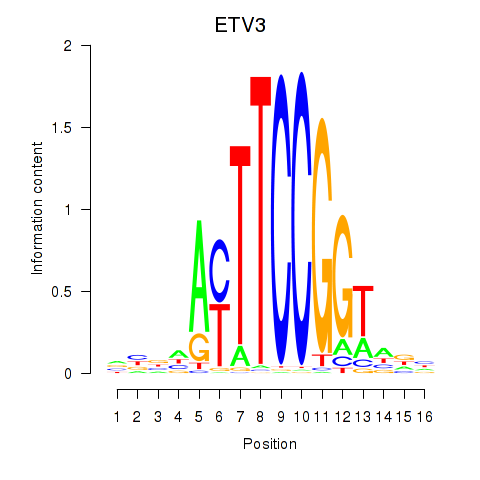
Results for ETV3
Z-value: 0.81
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETV3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV3 | hg19_v2_chr1_-_157108266_157108347 | 0.65 | 6.1e-03 | Click! |
Activity profile of ETV3 motif
Sorted Z-values of ETV3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_118846008 | 2.94 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr14_+_24702073 | 1.88 |
ENST00000399440.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_+_24702127 | 1.86 |
ENST00000557854.1 ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_+_24702099 | 1.83 |
ENST00000420554.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_+_24701628 | 1.82 |
ENST00000355299.4 ENST00000559836.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_+_24701819 | 1.81 |
ENST00000560139.1 ENST00000559910.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr11_-_61129335 | 1.81 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr17_-_56595196 | 1.44 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr2_+_32502952 | 1.41 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr5_-_130970723 | 1.24 |
ENST00000308008.6 ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr22_-_38245304 | 1.23 |
ENST00000609454.1 |
ANKRD54 |
ankyrin repeat domain 54 |
| chr2_+_138722028 | 1.12 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr20_-_48532019 | 1.07 |
ENST00000289431.5 |
SPATA2 |
spermatogenesis associated 2 |
| chr1_+_160313165 | 1.01 |
ENST00000421914.1 ENST00000535857.1 ENST00000438008.1 |
NCSTN |
nicastrin |
| chr17_-_39093672 | 1.01 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr20_-_34252847 | 1.01 |
ENST00000317619.3 ENST00000397446.1 ENST00000397445.1 ENST00000397443.1 ENST00000430570.1 ENST00000439806.2 ENST00000437340.1 ENST00000435161.1 ENST00000431148.1 |
CPNE1 RBM12 |
copine I RNA binding motif protein 12 |
| chr2_+_231729615 | 0.95 |
ENST00000326427.6 ENST00000335005.6 ENST00000326407.6 |
ITM2C |
integral membrane protein 2C |
| chr6_-_31620455 | 0.90 |
ENST00000437771.1 ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6 |
BCL2-associated athanogene 6 |
| chr6_-_31620403 | 0.90 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr11_-_62389449 | 0.89 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr3_-_15469006 | 0.83 |
ENST00000443029.1 ENST00000383790.3 ENST00000383789.5 |
METTL6 |
methyltransferase like 6 |
| chr22_+_47158578 | 0.80 |
ENST00000355704.3 |
TBC1D22A |
TBC1 domain family, member 22A |
| chr3_+_108308513 | 0.78 |
ENST00000361582.3 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr2_-_175260368 | 0.77 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chr1_+_160313062 | 0.75 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr12_+_132195617 | 0.72 |
ENST00000261674.4 ENST00000535236.1 ENST00000541286.1 |
SFSWAP |
splicing factor, suppressor of white-apricot homolog (Drosophila) |
| chr2_-_110371777 | 0.71 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr16_-_23652570 | 0.69 |
ENST00000261584.4 |
PALB2 |
partner and localizer of BRCA2 |
| chr15_+_74833518 | 0.68 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr22_+_38004832 | 0.67 |
ENST00000405147.3 ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr2_-_110371720 | 0.65 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr16_-_2097787 | 0.65 |
ENST00000566380.1 ENST00000219066.1 |
NTHL1 |
nth endonuclease III-like 1 (E. coli) |
| chr12_+_53693466 | 0.65 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr8_+_95565947 | 0.64 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr20_-_33999766 | 0.64 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr22_+_38004473 | 0.63 |
ENST00000414350.3 ENST00000343632.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr22_+_47158518 | 0.63 |
ENST00000337137.4 ENST00000380995.1 ENST00000407381.3 |
TBC1D22A |
TBC1 domain family, member 22A |
| chr12_+_53693812 | 0.62 |
ENST00000549488.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr16_+_8891670 | 0.60 |
ENST00000268261.4 ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2 |
phosphomannomutase 2 |
| chr18_-_72265035 | 0.60 |
ENST00000585279.1 ENST00000580048.1 |
LINC00909 |
long intergenic non-protein coding RNA 909 |
| chr17_+_37844331 | 0.59 |
ENST00000578199.1 ENST00000406381.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr12_-_6798616 | 0.58 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr11_+_46722368 | 0.58 |
ENST00000311764.2 |
ZNF408 |
zinc finger protein 408 |
| chr2_+_44589036 | 0.56 |
ENST00000402247.1 ENST00000407131.1 ENST00000403853.3 ENST00000378494.3 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
| chrX_+_47092314 | 0.56 |
ENST00000218348.3 |
USP11 |
ubiquitin specific peptidase 11 |
| chr1_+_11866207 | 0.56 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr13_-_46626847 | 0.56 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr1_-_92952433 | 0.56 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr17_-_7155274 | 0.56 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr21_+_44073916 | 0.56 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr16_+_70557685 | 0.56 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr2_+_138721850 | 0.56 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr17_-_7155802 | 0.55 |
ENST00000572043.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr3_+_15468862 | 0.54 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr2_-_9563575 | 0.53 |
ENST00000488451.1 ENST00000238091.4 ENST00000355346.4 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr1_+_156698234 | 0.51 |
ENST00000368218.4 ENST00000368216.4 |
RRNAD1 |
ribosomal RNA adenine dimethylase domain containing 1 |
| chr12_-_6798410 | 0.51 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr20_-_48532046 | 0.51 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr16_+_3493611 | 0.51 |
ENST00000407558.4 ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr1_+_168148273 | 0.51 |
ENST00000367830.3 |
TIPRL |
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr21_+_44073860 | 0.51 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chrX_-_74376108 | 0.50 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr16_+_14726672 | 0.50 |
ENST00000261658.2 ENST00000563971.1 |
BFAR |
bifunctional apoptosis regulator |
| chr15_-_72978490 | 0.50 |
ENST00000311755.3 |
HIGD2B |
HIG1 hypoxia inducible domain family, member 2B |
| chr14_+_35761580 | 0.50 |
ENST00000553809.1 ENST00000555764.1 ENST00000556506.1 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr7_+_99613212 | 0.48 |
ENST00000426572.1 ENST00000535170.1 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr8_+_117778736 | 0.48 |
ENST00000309822.2 ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23 |
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_20987982 | 0.47 |
ENST00000375048.3 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr12_+_6602517 | 0.47 |
ENST00000315579.5 ENST00000539714.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr14_-_74417096 | 0.47 |
ENST00000286544.3 |
FAM161B |
family with sequence similarity 161, member B |
| chr13_-_96705624 | 0.46 |
ENST00000376747.3 ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr14_+_35761540 | 0.46 |
ENST00000261479.4 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr12_-_6798523 | 0.46 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr16_+_2097970 | 0.45 |
ENST00000382538.6 ENST00000401874.2 ENST00000353929.4 |
TSC2 |
tuberous sclerosis 2 |
| chr17_-_29641084 | 0.45 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr15_-_90233866 | 0.44 |
ENST00000561257.1 |
PEX11A |
peroxisomal biogenesis factor 11 alpha |
| chr20_+_16710606 | 0.44 |
ENST00000377943.5 ENST00000246071.6 |
SNRPB2 |
small nuclear ribonucleoprotein polypeptide B |
| chr1_-_20987851 | 0.44 |
ENST00000464364.1 ENST00000602624.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr2_-_9563319 | 0.44 |
ENST00000497105.1 ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr9_-_86571628 | 0.43 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr20_-_2451395 | 0.43 |
ENST00000339610.6 ENST00000381342.2 ENST00000438552.2 |
SNRPB |
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr1_-_160001737 | 0.43 |
ENST00000368090.2 |
PIGM |
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr20_-_44993012 | 0.42 |
ENST00000372229.1 ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr5_+_14143728 | 0.42 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr15_-_90234006 | 0.42 |
ENST00000300056.3 ENST00000559170.1 |
PEX11A |
peroxisomal biogenesis factor 11 alpha |
| chr5_+_80597419 | 0.41 |
ENST00000254037.2 ENST00000407610.3 ENST00000380199.5 |
ZCCHC9 |
zinc finger, CCHC domain containing 9 |
| chr16_+_66968343 | 0.40 |
ENST00000417689.1 ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2 |
carboxylesterase 2 |
| chr17_-_29641104 | 0.40 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr14_-_24701539 | 0.40 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr1_+_11866270 | 0.40 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr16_-_89556942 | 0.40 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chr7_+_99613195 | 0.39 |
ENST00000324306.6 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr16_+_19535235 | 0.39 |
ENST00000565376.2 ENST00000396208.2 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chr19_+_56186606 | 0.39 |
ENST00000085079.7 |
EPN1 |
epsin 1 |
| chr17_-_34207295 | 0.39 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr16_-_23464459 | 0.39 |
ENST00000307149.5 |
COG7 |
component of oligomeric golgi complex 7 |
| chr9_-_26892765 | 0.39 |
ENST00000520187.1 ENST00000333916.5 |
CAAP1 |
caspase activity and apoptosis inhibitor 1 |
| chr20_+_62612470 | 0.39 |
ENST00000266079.4 ENST00000535781.1 |
PRPF6 |
pre-mRNA processing factor 6 |
| chr19_-_16653226 | 0.38 |
ENST00000198939.6 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr16_+_2098003 | 0.38 |
ENST00000439673.2 ENST00000350773.4 |
TSC2 |
tuberous sclerosis 2 |
| chr20_-_34287103 | 0.37 |
ENST00000374085.1 ENST00000419569.1 |
NFS1 |
NFS1 cysteine desulfurase |
| chr20_-_34638841 | 0.37 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chr16_-_11945370 | 0.37 |
ENST00000573251.1 ENST00000355674.5 ENST00000542106.1 ENST00000571133.1 |
RSL1D1 |
ribosomal L1 domain containing 1 |
| chr9_+_37753795 | 0.37 |
ENST00000377753.2 ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B |
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chr6_+_33239787 | 0.37 |
ENST00000439602.2 ENST00000474973.1 |
RPS18 |
ribosomal protein S18 |
| chr19_+_50381308 | 0.36 |
ENST00000599049.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr5_-_93447333 | 0.36 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr19_-_16653325 | 0.36 |
ENST00000546361.2 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr5_+_133984462 | 0.36 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr16_-_8962544 | 0.36 |
ENST00000570125.1 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr1_-_20987889 | 0.36 |
ENST00000415136.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr1_+_204485503 | 0.36 |
ENST00000367182.3 ENST00000507825.2 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
| chr11_+_47270436 | 0.35 |
ENST00000395397.3 ENST00000405576.1 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr6_+_28048753 | 0.35 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr17_-_37844267 | 0.35 |
ENST00000579146.1 ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3 |
post-GPI attachment to proteins 3 |
| chr22_+_41865109 | 0.35 |
ENST00000216254.4 ENST00000396512.3 |
ACO2 |
aconitase 2, mitochondrial |
| chr12_-_498620 | 0.35 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr16_+_19535133 | 0.34 |
ENST00000396212.2 ENST00000381396.5 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chr20_-_34287259 | 0.34 |
ENST00000397425.1 ENST00000540053.1 ENST00000541387.1 ENST00000374092.4 |
NFS1 |
NFS1 cysteine desulfurase |
| chr2_+_61404624 | 0.33 |
ENST00000394457.3 |
AHSA2 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr15_+_40331456 | 0.33 |
ENST00000504245.1 ENST00000560341.1 |
SRP14-AS1 |
SRP14 antisense RNA1 (head to head) |
| chr3_-_69101461 | 0.33 |
ENST00000543976.1 |
TMF1 |
TATA element modulatory factor 1 |
| chr14_-_24711806 | 0.33 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr2_+_201754050 | 0.33 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_-_56632592 | 0.33 |
ENST00000587279.1 ENST00000270459.3 |
ZNF787 |
zinc finger protein 787 |
| chr11_+_47270475 | 0.33 |
ENST00000481889.2 ENST00000436778.1 ENST00000531660.1 ENST00000407404.1 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr2_+_201754135 | 0.33 |
ENST00000409357.1 ENST00000409129.2 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr16_-_8962200 | 0.33 |
ENST00000562843.1 ENST00000561530.1 ENST00000396593.2 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chrX_+_51546103 | 0.32 |
ENST00000375772.3 |
MAGED1 |
melanoma antigen family D, 1 |
| chr14_-_24711865 | 0.32 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr16_-_31519691 | 0.32 |
ENST00000567994.1 ENST00000430477.2 ENST00000570164.1 ENST00000327237.2 |
C16orf58 |
chromosome 16 open reading frame 58 |
| chr19_-_19774473 | 0.32 |
ENST00000357324.6 |
ATP13A1 |
ATPase type 13A1 |
| chr9_+_115983808 | 0.32 |
ENST00000374210.6 ENST00000374212.4 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
| chr2_+_162016916 | 0.32 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr11_-_73882029 | 0.32 |
ENST00000539061.1 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr6_-_41040268 | 0.31 |
ENST00000373154.2 ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr21_-_46707793 | 0.31 |
ENST00000331343.7 ENST00000349485.5 |
POFUT2 |
protein O-fucosyltransferase 2 |
| chr3_-_69101413 | 0.31 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chr22_+_38245414 | 0.31 |
ENST00000381683.6 ENST00000414316.1 ENST00000406934.1 ENST00000451427.1 |
EIF3L |
eukaryotic translation initiation factor 3, subunit L |
| chr2_+_170440902 | 0.31 |
ENST00000448752.2 ENST00000418888.1 ENST00000414307.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr7_+_108210012 | 0.31 |
ENST00000249356.3 |
DNAJB9 |
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr5_-_114961858 | 0.31 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr4_-_83812402 | 0.30 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr17_-_33905521 | 0.30 |
ENST00000225873.4 |
PEX12 |
peroxisomal biogenesis factor 12 |
| chr11_+_67250490 | 0.30 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr6_-_159421198 | 0.30 |
ENST00000252655.1 ENST00000297262.3 ENST00000367069.2 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
| chr11_-_61197480 | 0.30 |
ENST00000439958.3 ENST00000394888.4 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_170440844 | 0.29 |
ENST00000260970.3 ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr2_-_43823093 | 0.29 |
ENST00000405006.4 |
THADA |
thyroid adenoma associated |
| chr8_-_53626974 | 0.29 |
ENST00000435644.2 ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1 |
RB1-inducible coiled-coil 1 |
| chr6_-_43484718 | 0.29 |
ENST00000372422.2 |
YIPF3 |
Yip1 domain family, member 3 |
| chr17_+_4843413 | 0.29 |
ENST00000572430.1 ENST00000262482.6 |
RNF167 |
ring finger protein 167 |
| chr19_+_11546440 | 0.29 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr6_-_41040195 | 0.29 |
ENST00000463088.1 ENST00000469104.1 ENST00000486443.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr5_-_74062930 | 0.29 |
ENST00000509430.1 ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2 |
G elongation factor, mitochondrial 2 |
| chr11_-_61197187 | 0.29 |
ENST00000449811.1 ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_162016804 | 0.29 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr4_-_83812248 | 0.29 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr17_+_27055798 | 0.28 |
ENST00000268766.6 |
NEK8 |
NIMA-related kinase 8 |
| chr19_+_42363917 | 0.28 |
ENST00000598742.1 |
RPS19 |
ribosomal protein S19 |
| chr1_-_173174681 | 0.28 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr15_+_77287426 | 0.28 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr19_+_56186557 | 0.28 |
ENST00000270460.6 |
EPN1 |
epsin 1 |
| chr4_+_40198527 | 0.28 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr17_+_57784826 | 0.27 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr2_-_43823119 | 0.27 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr6_-_43484621 | 0.27 |
ENST00000506469.1 ENST00000503972.1 |
YIPF3 |
Yip1 domain family, member 3 |
| chr22_+_30752606 | 0.27 |
ENST00000399824.2 ENST00000405659.1 ENST00000338306.3 |
CCDC157 |
coiled-coil domain containing 157 |
| chr16_+_47495225 | 0.27 |
ENST00000299167.8 ENST00000323584.5 ENST00000563376.1 |
PHKB |
phosphorylase kinase, beta |
| chr17_+_7387677 | 0.26 |
ENST00000322644.6 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr2_-_175351744 | 0.26 |
ENST00000295500.4 ENST00000392552.2 ENST00000392551.2 |
GPR155 |
G protein-coupled receptor 155 |
| chr11_-_71791435 | 0.25 |
ENST00000351960.6 ENST00000541719.1 ENST00000535111.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr14_+_69865401 | 0.25 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr2_+_70121075 | 0.25 |
ENST00000409116.1 |
SNRNP27 |
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr17_-_74722536 | 0.25 |
ENST00000585429.1 |
JMJD6 |
jumonji domain containing 6 |
| chr19_-_9929708 | 0.25 |
ENST00000247977.4 ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
| chr19_+_47759716 | 0.25 |
ENST00000221922.6 |
CCDC9 |
coiled-coil domain containing 9 |
| chr17_+_4843654 | 0.25 |
ENST00000575111.1 |
RNF167 |
ring finger protein 167 |
| chr2_+_128848881 | 0.24 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr19_-_55690758 | 0.24 |
ENST00000590851.1 |
SYT5 |
synaptotagmin V |
| chr19_+_13056663 | 0.24 |
ENST00000541222.1 ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
| chr12_+_108908962 | 0.24 |
ENST00000552695.1 ENST00000552758.1 ENST00000361549.2 |
FICD |
FIC domain containing |
| chr2_-_69870747 | 0.24 |
ENST00000409068.1 |
AAK1 |
AP2 associated kinase 1 |
| chr17_-_72869140 | 0.24 |
ENST00000583917.1 ENST00000293195.5 ENST00000442102.2 |
FDXR |
ferredoxin reductase |
| chr19_+_48248779 | 0.24 |
ENST00000246802.5 |
GLTSCR2 |
glioma tumor suppressor candidate region gene 2 |
| chr17_-_56065484 | 0.23 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr17_+_41561317 | 0.23 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr13_+_50070491 | 0.23 |
ENST00000496612.1 ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11 |
PHD finger protein 11 |
| chr6_-_159420780 | 0.23 |
ENST00000449822.1 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
| chr15_-_65809991 | 0.23 |
ENST00000559526.1 ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr14_-_21979428 | 0.23 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr12_-_7079805 | 0.23 |
ENST00000536316.2 ENST00000542912.1 ENST00000440277.1 ENST00000545167.1 ENST00000546111.1 ENST00000399433.2 ENST00000535923.1 |
PHB2 |
prohibitin 2 |
| chr17_+_4843679 | 0.23 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr13_-_45915221 | 0.23 |
ENST00000309246.5 ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1 |
tumor protein, translationally-controlled 1 |
| chr1_-_235292250 | 0.23 |
ENST00000366607.4 |
TOMM20 |
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr11_-_46722117 | 0.23 |
ENST00000311956.4 |
ARHGAP1 |
Rho GTPase activating protein 1 |
| chr18_+_5238549 | 0.22 |
ENST00000580684.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr18_-_47813940 | 0.22 |
ENST00000586837.1 ENST00000412036.2 ENST00000589940.1 |
CXXC1 |
CXXC finger protein 1 |
| chr11_-_71791518 | 0.22 |
ENST00000537217.1 ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr5_-_132202329 | 0.22 |
ENST00000378673.2 |
GDF9 |
growth differentiation factor 9 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.7 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 0.6 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 0.6 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.2 | 0.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 2.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 1.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.4 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:1904379 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.3 | 9.2 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.3 | 1.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 2.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 0.7 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.2 | 0.7 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.2 | 0.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.7 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 0.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.6 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 0.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.6 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.6 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.6 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.5 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.5 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 1.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.8 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 1.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.8 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.4 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.5 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 1.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.2 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.1 | GO:0071392 | cellular response to estrogen stimulus(GO:0071391) cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 1.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.3 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0097480 | synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.7 | 2.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.8 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.4 | 1.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 0.7 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 0.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.9 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.2 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 1.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |