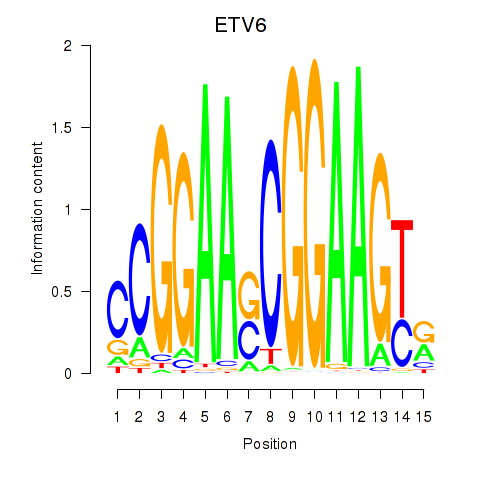
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ETV6
Z-value: 0.97
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETV6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV6 | hg19_v2_chr12_+_11802753_11802834 | 0.30 | 2.6e-01 | Click! |
Activity profile of ETV6 motif
Sorted Z-values of ETV6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_31620191 | 1.55 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr2_+_201981527 | 0.98 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr12_+_120105558 | 0.92 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr19_-_9731872 | 0.88 |
ENST00000424629.1 ENST00000326044.5 ENST00000354661.4 ENST00000435550.1 ENST00000444611.1 ENST00000421525.1 |
ZNF561 |
zinc finger protein 561 |
| chr2_-_96971259 | 0.85 |
ENST00000349783.5 |
SNRNP200 |
small nuclear ribonucleoprotein 200kDa (U5) |
| chr1_+_26644441 | 0.82 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr11_-_61129335 | 0.79 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr2_+_201981119 | 0.74 |
ENST00000395148.2 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr5_+_102455968 | 0.73 |
ENST00000358359.3 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr13_-_46679185 | 0.69 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr21_-_46340884 | 0.67 |
ENST00000302347.5 ENST00000517819.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr13_-_46679144 | 0.67 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr9_-_33264557 | 0.66 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr20_-_3140490 | 0.65 |
ENST00000449731.1 ENST00000380266.3 |
UBOX5 FASTKD5 |
U-box domain containing 5 FAST kinase domains 5 |
| chr2_-_96971232 | 0.63 |
ENST00000323853.5 |
SNRNP200 |
small nuclear ribonucleoprotein 200kDa (U5) |
| chr6_+_30525051 | 0.63 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr6_-_31697255 | 0.63 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_33264676 | 0.61 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr21_-_46340770 | 0.61 |
ENST00000397854.3 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_+_30524663 | 0.59 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr17_-_38574169 | 0.58 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr6_+_17600576 | 0.58 |
ENST00000259963.3 |
FAM8A1 |
family with sequence similarity 8, member A1 |
| chr15_+_75074410 | 0.56 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr5_+_102455853 | 0.55 |
ENST00000515845.1 ENST00000321521.9 ENST00000507921.1 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr22_+_37309662 | 0.55 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr6_-_31697563 | 0.53 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chrX_+_69509927 | 0.51 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr6_-_31697977 | 0.50 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr17_+_80416482 | 0.49 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chr20_-_20033052 | 0.49 |
ENST00000536226.1 |
CRNKL1 |
crooked neck pre-mRNA splicing factor 1 |
| chr7_-_148823387 | 0.48 |
ENST00000483014.1 ENST00000378061.2 |
ZNF425 |
zinc finger protein 425 |
| chr17_+_79935418 | 0.48 |
ENST00000306729.7 ENST00000306739.4 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr19_-_9546227 | 0.48 |
ENST00000361451.2 ENST00000361151.1 |
ZNF266 |
zinc finger protein 266 |
| chr2_-_85839146 | 0.47 |
ENST00000306336.5 ENST00000409734.3 |
C2orf68 |
chromosome 2 open reading frame 68 |
| chr17_+_79935464 | 0.46 |
ENST00000581647.1 ENST00000580534.1 ENST00000579684.1 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_-_62389449 | 0.46 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr6_-_31620403 | 0.44 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr17_-_56429500 | 0.44 |
ENST00000225504.3 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
| chr6_-_31620149 | 0.44 |
ENST00000435080.1 ENST00000375976.4 ENST00000441054.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr11_-_67205538 | 0.43 |
ENST00000326294.3 |
PTPRCAP |
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr19_-_9929484 | 0.42 |
ENST00000586651.1 ENST00000586073.1 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
| chr17_+_80416050 | 0.42 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chrX_+_47092314 | 0.41 |
ENST00000218348.3 |
USP11 |
ubiquitin specific peptidase 11 |
| chr6_-_31620455 | 0.41 |
ENST00000437771.1 ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6 |
BCL2-associated athanogene 6 |
| chr19_+_6887571 | 0.40 |
ENST00000250572.8 ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr2_-_75938115 | 0.40 |
ENST00000321027.3 |
GCFC2 |
GC-rich sequence DNA-binding factor 2 |
| chr14_+_100531615 | 0.40 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr19_-_9929708 | 0.40 |
ENST00000247977.4 ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
| chr20_+_16710606 | 0.40 |
ENST00000377943.5 ENST00000246071.6 |
SNRPB2 |
small nuclear ribonucleoprotein polypeptide B |
| chr19_-_9546177 | 0.40 |
ENST00000592292.1 ENST00000588221.1 |
ZNF266 |
zinc finger protein 266 |
| chr12_+_124196865 | 0.40 |
ENST00000330342.3 |
ATP6V0A2 |
ATPase, H+ transporting, lysosomal V0 subunit a2 |
| chr17_-_76836963 | 0.39 |
ENST00000312010.6 |
USP36 |
ubiquitin specific peptidase 36 |
| chr2_-_128615681 | 0.38 |
ENST00000409955.1 ENST00000272645.4 |
POLR2D |
polymerase (RNA) II (DNA directed) polypeptide D |
| chr19_+_57078854 | 0.38 |
ENST00000330619.8 ENST00000391709.3 ENST00000601902.1 |
ZNF470 |
zinc finger protein 470 |
| chr2_-_101034070 | 0.38 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr19_+_50919056 | 0.38 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr6_-_11382478 | 0.37 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr5_+_36152091 | 0.37 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_-_225907150 | 0.37 |
ENST00000258390.7 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr2_-_75937994 | 0.37 |
ENST00000409857.3 ENST00000470503.1 ENST00000541687.1 ENST00000442309.1 |
GCFC2 |
GC-rich sequence DNA-binding factor 2 |
| chr5_-_40755987 | 0.37 |
ENST00000337702.4 |
TTC33 |
tetratricopeptide repeat domain 33 |
| chr16_+_67226019 | 0.36 |
ENST00000379378.3 |
E2F4 |
E2F transcription factor 4, p107/p130-binding |
| chrX_-_100604184 | 0.36 |
ENST00000372902.3 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr11_+_47600562 | 0.35 |
ENST00000263774.4 ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3 |
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr15_+_91427691 | 0.35 |
ENST00000559355.1 ENST00000394302.1 |
FES |
feline sarcoma oncogene |
| chr1_-_150207017 | 0.35 |
ENST00000369119.3 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_126307576 | 0.34 |
ENST00000334379.5 ENST00000450358.1 ENST00000368332.3 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr1_+_22351977 | 0.34 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr16_+_70557685 | 0.34 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr9_-_135282195 | 0.34 |
ENST00000334270.2 |
TTF1 |
transcription termination factor, RNA polymerase I |
| chr17_-_33905521 | 0.34 |
ENST00000225873.4 |
PEX12 |
peroxisomal biogenesis factor 12 |
| chr9_-_115480303 | 0.33 |
ENST00000374234.1 ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP |
INTS3 and NABP interacting protein |
| chr15_+_91427642 | 0.33 |
ENST00000328850.3 ENST00000414248.2 |
FES |
feline sarcoma oncogene |
| chr17_+_75137460 | 0.32 |
ENST00000587820.1 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr19_-_45681482 | 0.32 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr20_+_3190006 | 0.32 |
ENST00000380113.3 ENST00000455664.2 ENST00000399838.3 |
ITPA |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr15_+_75074385 | 0.31 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr17_+_7487146 | 0.31 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr19_+_7069690 | 0.31 |
ENST00000439035.2 |
ZNF557 |
zinc finger protein 557 |
| chr16_+_3493611 | 0.31 |
ENST00000407558.4 ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chrX_+_48542168 | 0.31 |
ENST00000376701.4 |
WAS |
Wiskott-Aldrich syndrome |
| chr17_-_56595196 | 0.31 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr15_+_41624892 | 0.30 |
ENST00000260359.6 ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1 |
nucleolar and spindle associated protein 1 |
| chr16_+_10479906 | 0.30 |
ENST00000562527.1 ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2 |
activating transcription factor 7 interacting protein 2 |
| chr2_+_128848740 | 0.30 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr10_+_99079008 | 0.30 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr3_-_49142178 | 0.29 |
ENST00000452739.1 ENST00000414533.1 ENST00000417025.1 |
QARS |
glutaminyl-tRNA synthetase |
| chr2_+_122494676 | 0.29 |
ENST00000455432.1 |
TSN |
translin |
| chr16_+_10837643 | 0.29 |
ENST00000574334.1 ENST00000283027.5 ENST00000433392.2 |
NUBP1 |
nucleotide binding protein 1 |
| chr19_+_7069426 | 0.29 |
ENST00000252840.6 ENST00000414706.1 |
ZNF557 |
zinc finger protein 557 |
| chr14_-_106692191 | 0.29 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr5_-_137090028 | 0.28 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr19_+_14800711 | 0.28 |
ENST00000536363.1 ENST00000540689.2 ENST00000601134.1 ENST00000292530.6 |
ZNF333 |
zinc finger protein 333 |
| chr6_+_170102210 | 0.28 |
ENST00000439249.1 ENST00000332290.2 |
C6orf120 |
chromosome 6 open reading frame 120 |
| chr19_-_54618650 | 0.28 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr10_+_35415719 | 0.28 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chrX_+_110924346 | 0.28 |
ENST00000371979.3 ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13 |
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr1_+_39456895 | 0.28 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr9_+_131267052 | 0.27 |
ENST00000539582.1 |
GLE1 |
GLE1 RNA export mediator |
| chr14_-_24616426 | 0.27 |
ENST00000216802.5 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr15_-_80263506 | 0.27 |
ENST00000335661.6 |
BCL2A1 |
BCL2-related protein A1 |
| chr17_-_76836729 | 0.27 |
ENST00000587783.1 ENST00000542802.3 ENST00000586531.1 ENST00000589424.1 ENST00000590546.2 |
USP36 |
ubiquitin specific peptidase 36 |
| chr17_+_76374714 | 0.27 |
ENST00000262764.6 ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
| chr2_+_241544834 | 0.26 |
ENST00000319838.5 ENST00000403859.1 ENST00000438013.2 |
GPR35 |
G protein-coupled receptor 35 |
| chr3_-_49142504 | 0.26 |
ENST00000306125.6 ENST00000420147.2 |
QARS |
glutaminyl-tRNA synthetase |
| chr1_-_165738072 | 0.26 |
ENST00000481278.1 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr17_+_72744791 | 0.26 |
ENST00000583369.1 ENST00000262613.5 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
| chr19_+_55897699 | 0.26 |
ENST00000558131.1 ENST00000558752.1 ENST00000458349.2 |
RPL28 |
ribosomal protein L28 |
| chr15_+_74833518 | 0.25 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr6_+_26156551 | 0.25 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr12_-_57914275 | 0.25 |
ENST00000547303.1 ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3 |
DNA-damage-inducible transcript 3 |
| chr7_-_99679324 | 0.25 |
ENST00000292393.5 ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3 |
zinc finger protein 3 |
| chr11_+_65770227 | 0.25 |
ENST00000527348.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr17_+_26989109 | 0.25 |
ENST00000314616.6 ENST00000347486.4 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr6_+_42896865 | 0.24 |
ENST00000372836.4 ENST00000394142.3 |
CNPY3 |
canopy FGF signaling regulator 3 |
| chr1_-_114301755 | 0.24 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr5_+_36152163 | 0.24 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr22_+_40742497 | 0.24 |
ENST00000216194.7 |
ADSL |
adenylosuccinate lyase |
| chr22_+_40742512 | 0.24 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr5_+_36152179 | 0.24 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr19_+_54619125 | 0.23 |
ENST00000445811.1 ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31 |
pre-mRNA processing factor 31 |
| chr15_+_91260552 | 0.23 |
ENST00000355112.3 ENST00000560509.1 |
BLM |
Bloom syndrome, RecQ helicase-like |
| chr5_+_31532373 | 0.23 |
ENST00000325366.9 ENST00000355907.3 ENST00000507818.2 |
C5orf22 |
chromosome 5 open reading frame 22 |
| chr7_-_37488834 | 0.23 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr2_+_128848881 | 0.23 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_+_43380459 | 0.23 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr2_+_85839218 | 0.22 |
ENST00000448971.1 ENST00000442708.1 ENST00000450066.2 |
USP39 |
ubiquitin specific peptidase 39 |
| chr12_-_48551366 | 0.22 |
ENST00000535988.1 ENST00000536953.1 ENST00000535055.1 ENST00000317697.3 ENST00000536549.1 |
ASB8 |
ankyrin repeat and SOCS box containing 8 |
| chr3_-_196669298 | 0.22 |
ENST00000411704.1 ENST00000452404.2 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
| chr17_+_61904766 | 0.22 |
ENST00000581842.1 ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr19_-_56826157 | 0.22 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr19_+_49375649 | 0.21 |
ENST00000200453.5 |
PPP1R15A |
protein phosphatase 1, regulatory subunit 15A |
| chr11_-_60674037 | 0.21 |
ENST00000541371.1 ENST00000227524.4 |
PRPF19 |
pre-mRNA processing factor 19 |
| chr17_+_34958001 | 0.21 |
ENST00000250156.7 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr6_+_57182400 | 0.21 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr16_-_70557430 | 0.21 |
ENST00000393612.4 ENST00000564653.1 ENST00000323786.5 |
COG4 |
component of oligomeric golgi complex 4 |
| chr2_-_136743169 | 0.20 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr1_+_95699740 | 0.20 |
ENST00000429514.2 ENST00000263893.6 |
RWDD3 |
RWD domain containing 3 |
| chr19_-_6393465 | 0.20 |
ENST00000394456.5 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr14_+_24616588 | 0.20 |
ENST00000324103.6 ENST00000559260.1 |
RNF31 |
ring finger protein 31 |
| chr12_+_64845660 | 0.20 |
ENST00000331710.5 |
TBK1 |
TANK-binding kinase 1 |
| chr12_-_76742183 | 0.20 |
ENST00000393262.3 |
BBS10 |
Bardet-Biedl syndrome 10 |
| chr17_+_60501228 | 0.20 |
ENST00000311506.5 |
METTL2A |
methyltransferase like 2A |
| chr3_-_52740012 | 0.20 |
ENST00000407584.3 ENST00000266014.5 |
GLT8D1 |
glycosyltransferase 8 domain containing 1 |
| chr11_+_65769946 | 0.20 |
ENST00000533166.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr14_+_21458127 | 0.20 |
ENST00000382985.4 ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17 |
methyltransferase like 17 |
| chr11_+_65769550 | 0.20 |
ENST00000312175.2 ENST00000445560.2 ENST00000530204.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr5_-_31532160 | 0.20 |
ENST00000511367.2 ENST00000513349.1 |
DROSHA |
drosha, ribonuclease type III |
| chr1_+_236958554 | 0.20 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr9_+_37753795 | 0.19 |
ENST00000377753.2 ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B |
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chr15_-_85259360 | 0.19 |
ENST00000559729.1 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
| chr19_-_54619006 | 0.19 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr9_+_72873837 | 0.19 |
ENST00000361138.5 |
SMC5 |
structural maintenance of chromosomes 5 |
| chr19_+_19303008 | 0.19 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr9_-_139268068 | 0.19 |
ENST00000371734.3 ENST00000371732.5 ENST00000315908.7 |
CARD9 |
caspase recruitment domain family, member 9 |
| chr10_+_43278217 | 0.19 |
ENST00000374518.5 |
BMS1 |
BMS1 ribosome biogenesis factor |
| chr19_-_6393216 | 0.18 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr16_+_3507985 | 0.18 |
ENST00000421765.3 ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr6_-_13814663 | 0.18 |
ENST00000359495.2 ENST00000379170.4 |
MCUR1 |
mitochondrial calcium uniporter regulator 1 |
| chrX_+_46306624 | 0.18 |
ENST00000360017.5 |
KRBOX4 |
KRAB box domain containing 4 |
| chr16_+_19535235 | 0.17 |
ENST00000565376.2 ENST00000396208.2 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chr19_-_14016877 | 0.17 |
ENST00000454313.1 ENST00000591586.1 ENST00000346736.2 |
C19orf57 |
chromosome 19 open reading frame 57 |
| chr11_+_60681346 | 0.17 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr8_+_117778736 | 0.17 |
ENST00000309822.2 ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23 |
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr2_-_201828356 | 0.17 |
ENST00000234296.2 |
ORC2 |
origin recognition complex, subunit 2 |
| chr5_+_179247759 | 0.17 |
ENST00000389805.4 ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1 |
sequestosome 1 |
| chr17_-_73975444 | 0.17 |
ENST00000293217.5 ENST00000537812.1 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
| chr16_+_3508063 | 0.17 |
ENST00000576787.1 ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chrX_+_1710484 | 0.17 |
ENST00000313871.3 ENST00000381261.3 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
| chr11_+_65408273 | 0.17 |
ENST00000394227.3 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr4_-_74124502 | 0.16 |
ENST00000358602.4 ENST00000330838.6 ENST00000561029.1 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr4_+_1723197 | 0.16 |
ENST00000485989.2 ENST00000313288.4 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
| chr5_-_37371163 | 0.16 |
ENST00000513532.1 |
NUP155 |
nucleoporin 155kDa |
| chr7_-_128694927 | 0.16 |
ENST00000471166.1 ENST00000265388.5 |
TNPO3 |
transportin 3 |
| chr12_-_125348329 | 0.16 |
ENST00000546215.1 ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr20_-_34542548 | 0.16 |
ENST00000305978.2 |
SCAND1 |
SCAN domain containing 1 |
| chr16_+_19535133 | 0.16 |
ENST00000396212.2 ENST00000381396.5 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chrX_+_122994112 | 0.16 |
ENST00000355640.3 |
XIAP |
X-linked inhibitor of apoptosis |
| chr4_-_8442438 | 0.16 |
ENST00000356406.5 ENST00000413009.2 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
| chr9_+_100818976 | 0.16 |
ENST00000210444.5 |
NANS |
N-acetylneuraminic acid synthase |
| chr1_+_27648709 | 0.16 |
ENST00000608611.1 ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222 |
transmembrane protein 222 |
| chr7_-_15726296 | 0.16 |
ENST00000262041.5 |
MEOX2 |
mesenchyme homeobox 2 |
| chr19_+_52772832 | 0.16 |
ENST00000593703.1 ENST00000601711.1 ENST00000599581.1 |
ZNF766 |
zinc finger protein 766 |
| chr15_+_74908147 | 0.16 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr8_-_48872686 | 0.15 |
ENST00000314191.2 ENST00000338368.3 |
PRKDC |
protein kinase, DNA-activated, catalytic polypeptide |
| chr3_+_9404526 | 0.15 |
ENST00000452837.2 ENST00000417036.1 ENST00000419437.1 ENST00000345094.3 ENST00000515662.2 |
THUMPD3 |
THUMP domain containing 3 |
| chr1_+_27648648 | 0.15 |
ENST00000374076.4 |
TMEM222 |
transmembrane protein 222 |
| chr14_+_100842735 | 0.15 |
ENST00000554998.1 ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25 |
WD repeat domain 25 |
| chr9_-_80263220 | 0.15 |
ENST00000341700.6 |
GNA14 |
guanine nucleotide binding protein (G protein), alpha 14 |
| chr12_+_10658201 | 0.15 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr17_-_79876010 | 0.15 |
ENST00000328666.6 |
SIRT7 |
sirtuin 7 |
| chr5_-_37371278 | 0.15 |
ENST00000231498.3 |
NUP155 |
nucleoporin 155kDa |
| chr16_+_23652700 | 0.15 |
ENST00000300087.2 |
DCTN5 |
dynactin 5 (p25) |
| chr16_-_67693846 | 0.14 |
ENST00000602850.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr16_+_48278178 | 0.14 |
ENST00000285737.4 ENST00000535754.1 |
LONP2 |
lon peptidase 2, peroxisomal |
| chr3_-_196669248 | 0.14 |
ENST00000447325.1 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
| chr12_-_121734489 | 0.14 |
ENST00000412367.2 ENST00000402834.4 ENST00000404169.3 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr15_-_85259294 | 0.14 |
ENST00000558217.1 ENST00000558196.1 ENST00000558134.1 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
| chr17_-_37844267 | 0.14 |
ENST00000579146.1 ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3 |
post-GPI attachment to proteins 3 |
| chr15_-_85259384 | 0.14 |
ENST00000455959.3 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.6 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 1.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.5 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 1.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.6 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.4 | 1.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.3 | 1.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 1.3 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 0.6 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.2 | 0.5 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 0.5 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.2 | 0.9 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 1.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.3 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 1.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.2 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 1.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.6 | GO:0030263 | resolution of meiotic recombination intermediates(GO:0000712) apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.7 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 1.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.4 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0032632 | response to molecule of fungal origin(GO:0002238) interleukin-3 production(GO:0032632) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.5 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.1 | GO:0071569 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) protein ufmylation(GO:0071569) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |