Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
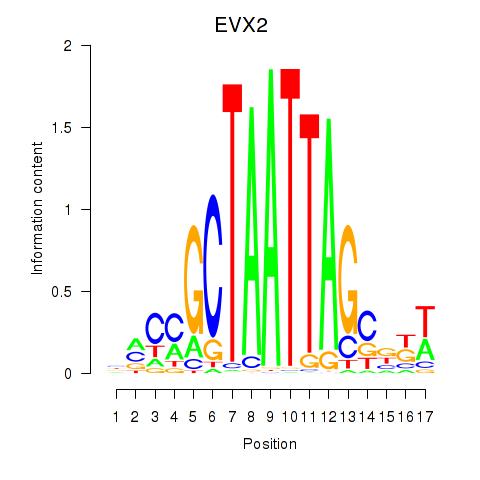
Results for EVX2
Z-value: 0.61
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | EVX2 |
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_55562479 | 1.95 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 1.87 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55562582 | 1.34 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr11_-_115375107 | 0.90 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr4_-_41884620 | 0.69 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr15_+_41057818 | 0.60 |
ENST00000558467.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr3_+_111718173 | 0.49 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr12_+_28410128 | 0.46 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr3_+_111718036 | 0.30 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr17_-_27418537 | 0.25 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_+_28261533 | 0.25 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr6_-_111927062 | 0.24 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr3_+_111717600 | 0.23 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr3_+_111717511 | 0.20 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr17_-_40337470 | 0.20 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr17_-_9929581 | 0.19 |
ENST00000437099.2 ENST00000396115.2 |
GAS7 |
growth arrest-specific 7 |
| chr18_-_33709268 | 0.11 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr1_+_28261492 | 0.10 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_28261621 | 0.09 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_160160346 | 0.08 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_+_115342349 | 0.07 |
ENST00000393780.3 |
GAP43 |
growth associated protein 43 |
| chr1_-_151431647 | 0.06 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr6_-_62996066 | 0.01 |
ENST00000281156.4 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.6 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.2 | 5.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |