Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
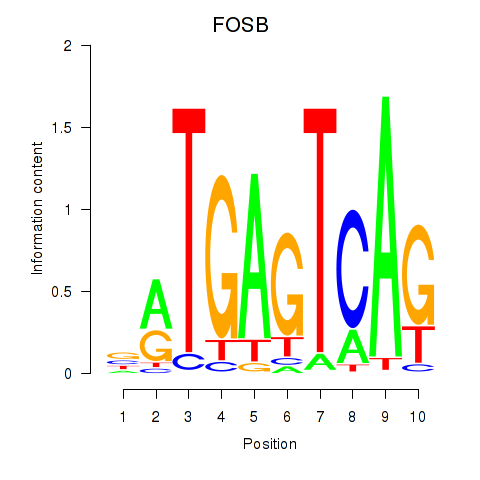
Results for FOSB
Z-value: 1.31
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.9 | FOSB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSB | hg19_v2_chr19_+_45973120_45973171, hg19_v2_chr19_+_45971246_45971265 | 0.06 | 8.3e-01 | Click! |
Activity profile of FOSB motif
Sorted Z-values of FOSB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21452804 | 10.13 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452964 | 9.84 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_61143994 | 4.92 |
ENST00000382771.4 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr2_+_102618428 | 3.81 |
ENST00000457817.1 |
IL1R2 |
interleukin 1 receptor, type II |
| chr17_-_39769005 | 3.70 |
ENST00000301653.4 ENST00000593067.1 |
KRT16 |
keratin 16 |
| chr1_+_27189631 | 3.14 |
ENST00000339276.4 |
SFN |
stratifin |
| chr19_-_39264072 | 2.99 |
ENST00000599035.1 ENST00000378626.4 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
| chr6_+_125540951 | 2.88 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr3_-_151034734 | 2.71 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr18_+_61442629 | 2.45 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr11_+_394196 | 2.36 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr1_+_152881014 | 2.35 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr17_-_39942940 | 2.24 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr17_+_73717407 | 2.09 |
ENST00000579662.1 |
ITGB4 |
integrin, beta 4 |
| chr17_+_73717551 | 1.99 |
ENST00000450894.3 |
ITGB4 |
integrin, beta 4 |
| chrX_-_154563889 | 1.94 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr12_-_15114603 | 1.82 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_26869597 | 1.77 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr12_-_15114492 | 1.75 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_-_36246060 | 1.75 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_+_73717516 | 1.57 |
ENST00000200181.3 ENST00000339591.3 |
ITGB4 |
integrin, beta 4 |
| chr6_+_106959718 | 1.50 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chr16_+_30751953 | 1.23 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr17_-_38721711 | 1.18 |
ENST00000578085.1 ENST00000246657.2 |
CCR7 |
chemokine (C-C motif) receptor 7 |
| chr11_-_108408895 | 1.14 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chr1_-_24126023 | 1.14 |
ENST00000429356.1 |
GALE |
UDP-galactose-4-epimerase |
| chr3_-_48632593 | 1.13 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr3_+_36421826 | 1.12 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr1_-_95007193 | 1.09 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr6_-_35888905 | 1.08 |
ENST00000510290.1 ENST00000423325.2 ENST00000373822.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr1_-_153029980 | 1.07 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr17_+_30771279 | 0.99 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr6_+_292051 | 0.99 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr5_-_78809950 | 0.99 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr17_+_74381343 | 0.92 |
ENST00000392496.3 |
SPHK1 |
sphingosine kinase 1 |
| chr8_-_42623747 | 0.91 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr6_-_35888824 | 0.91 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr15_+_40531243 | 0.90 |
ENST00000558055.1 ENST00000455577.2 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr7_+_55177416 | 0.89 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr1_-_6662919 | 0.86 |
ENST00000377658.4 ENST00000377663.3 |
KLHL21 |
kelch-like family member 21 |
| chr1_-_207224307 | 0.86 |
ENST00000315927.4 |
YOD1 |
YOD1 deubiquitinase |
| chr22_+_23134974 | 0.85 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr15_+_40531621 | 0.82 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr8_-_70745575 | 0.82 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr2_-_85641162 | 0.81 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr11_+_59522837 | 0.80 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr17_-_29641104 | 0.79 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr14_-_23426337 | 0.75 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr3_+_157828152 | 0.73 |
ENST00000476899.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chr3_-_141747950 | 0.73 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_-_23426322 | 0.73 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr3_-_141747439 | 0.73 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_-_23426270 | 0.70 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr9_+_137218362 | 0.70 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr12_-_102591604 | 0.68 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr1_+_165796753 | 0.66 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr17_-_29641084 | 0.59 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr17_-_27405875 | 0.58 |
ENST00000359450.6 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_-_153521597 | 0.58 |
ENST00000368712.1 |
S100A3 |
S100 calcium binding protein A3 |
| chr3_-_141747459 | 0.57 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr15_+_75335604 | 0.56 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr5_-_175843524 | 0.54 |
ENST00000502877.1 |
CLTB |
clathrin, light chain B |
| chr8_-_42623924 | 0.53 |
ENST00000276410.2 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr5_+_66124590 | 0.53 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_108487245 | 0.51 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr2_-_238499303 | 0.51 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr5_+_139493665 | 0.48 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr3_-_113465065 | 0.48 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_+_103801140 | 0.46 |
ENST00000561325.1 ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chr11_-_118213455 | 0.46 |
ENST00000300692.4 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr17_-_73150629 | 0.45 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
| chr4_+_170581213 | 0.44 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr8_+_26150628 | 0.44 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_+_103035102 | 0.44 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr16_+_2083265 | 0.44 |
ENST00000565855.1 ENST00000566198.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr14_+_73525265 | 0.44 |
ENST00000525161.1 |
RBM25 |
RNA binding motif protein 25 |
| chr2_-_220173685 | 0.43 |
ENST00000423636.2 ENST00000442029.1 ENST00000412847.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr4_-_100356844 | 0.42 |
ENST00000437033.2 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_8765446 | 0.42 |
ENST00000537228.1 ENST00000229335.6 |
AICDA |
activation-induced cytidine deaminase |
| chr1_-_153521714 | 0.42 |
ENST00000368713.3 |
S100A3 |
S100 calcium binding protein A3 |
| chr14_-_96180435 | 0.41 |
ENST00000556450.1 ENST00000555202.1 ENST00000554012.1 ENST00000402399.1 |
TCL1A |
T-cell leukemia/lymphoma 1A |
| chr6_-_38670897 | 0.41 |
ENST00000373365.4 |
GLO1 |
glyoxalase I |
| chr19_+_926000 | 0.41 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr17_-_27503770 | 0.41 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr11_-_118213360 | 0.40 |
ENST00000529594.1 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr11_-_65667997 | 0.39 |
ENST00000312562.2 ENST00000534222.1 |
FOSL1 |
FOS-like antigen 1 |
| chr17_-_48785216 | 0.39 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr7_-_20256965 | 0.38 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr16_+_57662138 | 0.38 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr18_+_21693306 | 0.38 |
ENST00000540918.2 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr19_-_44259053 | 0.37 |
ENST00000601170.1 |
SMG9 |
SMG9 nonsense mediated mRNA decay factor |
| chr7_+_73624327 | 0.37 |
ENST00000361082.3 ENST00000275635.7 ENST00000470709.1 |
LAT2 |
linker for activation of T cells family, member 2 |
| chr20_+_44637526 | 0.37 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_+_44194762 | 0.36 |
ENST00000371708.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr9_-_124991124 | 0.36 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr12_-_53625958 | 0.36 |
ENST00000327550.3 ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG |
retinoic acid receptor, gamma |
| chr1_+_154975110 | 0.36 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr6_+_151042224 | 0.35 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr4_-_146019693 | 0.35 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr17_+_72426891 | 0.35 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_169926047 | 0.34 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr13_-_52027134 | 0.34 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr1_-_93257951 | 0.33 |
ENST00000543509.1 ENST00000370331.1 ENST00000540033.1 |
EVI5 |
ecotropic viral integration site 5 |
| chr4_-_100356291 | 0.32 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_-_44259136 | 0.31 |
ENST00000270066.6 |
SMG9 |
SMG9 nonsense mediated mRNA decay factor |
| chr17_-_72864739 | 0.31 |
ENST00000579893.1 ENST00000544854.1 |
FDXR |
ferredoxin reductase |
| chr20_+_21686290 | 0.31 |
ENST00000398485.2 |
PAX1 |
paired box 1 |
| chr16_+_57662419 | 0.31 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_-_65667884 | 0.31 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr1_+_154975258 | 0.30 |
ENST00000417934.2 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr11_-_66103867 | 0.30 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr14_+_103800513 | 0.29 |
ENST00000560338.1 ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chr2_-_65593784 | 0.29 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr11_-_2924970 | 0.29 |
ENST00000533594.1 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_-_49351303 | 0.29 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chr6_-_31704282 | 0.28 |
ENST00000375784.3 ENST00000375779.2 |
CLIC1 |
chloride intracellular channel 1 |
| chr7_+_112063192 | 0.27 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr15_+_101459420 | 0.26 |
ENST00000388948.3 ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1 |
leucine-rich repeat kinase 1 |
| chr11_-_47198380 | 0.26 |
ENST00000419701.2 ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2 |
ADP-ribosylation factor GTPase activating protein 2 |
| chr9_+_71819927 | 0.26 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr2_+_233562015 | 0.25 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr9_-_139948468 | 0.25 |
ENST00000312665.5 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr1_-_21948906 | 0.25 |
ENST00000374761.2 ENST00000599760.1 |
RAP1GAP |
RAP1 GTPase activating protein |
| chr7_+_87563557 | 0.25 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr19_-_51568324 | 0.25 |
ENST00000595547.1 ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13 |
kallikrein-related peptidase 13 |
| chr6_+_32121218 | 0.24 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chrX_+_41193407 | 0.24 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr4_-_100356551 | 0.24 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_124997766 | 0.23 |
ENST00000543970.1 |
RP11-83B20.1 |
RP11-83B20.1 |
| chr18_-_268019 | 0.23 |
ENST00000261600.6 |
THOC1 |
THO complex 1 |
| chr18_-_74207146 | 0.23 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chrX_-_153285251 | 0.22 |
ENST00000444230.1 ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
| chr1_+_44584522 | 0.22 |
ENST00000372299.3 |
KLF17 |
Kruppel-like factor 17 |
| chr1_-_220263096 | 0.22 |
ENST00000463953.1 ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1 |
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr16_-_72206034 | 0.22 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr10_-_29923893 | 0.22 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr12_-_71182695 | 0.21 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr11_+_128563652 | 0.21 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_59668550 | 0.21 |
ENST00000521764.1 |
NACA2 |
nascent polypeptide-associated complex alpha subunit 2 |
| chr12_-_118628350 | 0.21 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr1_-_236046872 | 0.21 |
ENST00000536965.1 |
LYST |
lysosomal trafficking regulator |
| chr10_+_88854926 | 0.20 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr9_+_71820057 | 0.20 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr10_+_85899196 | 0.19 |
ENST00000372134.3 |
GHITM |
growth hormone inducible transmembrane protein |
| chr11_+_10477733 | 0.19 |
ENST00000528723.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr2_-_11606275 | 0.18 |
ENST00000381525.3 ENST00000362009.4 |
E2F6 |
E2F transcription factor 6 |
| chr19_+_47104493 | 0.17 |
ENST00000291295.9 ENST00000597743.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr11_-_2924720 | 0.17 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_-_238499131 | 0.17 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr17_-_8021710 | 0.17 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr12_+_86268065 | 0.16 |
ENST00000551529.1 ENST00000256010.6 |
NTS |
neurotensin |
| chr11_-_66103932 | 0.16 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr1_+_101003687 | 0.16 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr2_-_136633940 | 0.16 |
ENST00000264156.2 |
MCM6 |
minichromosome maintenance complex component 6 |
| chr16_-_57935277 | 0.15 |
ENST00000565942.1 |
CNGB1 |
cyclic nucleotide gated channel beta 1 |
| chr1_+_223889310 | 0.15 |
ENST00000434648.1 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr2_-_219858123 | 0.14 |
ENST00000453769.1 ENST00000295728.2 ENST00000392096.2 |
CRYBA2 |
crystallin, beta A2 |
| chr7_-_14942283 | 0.13 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr3_+_183903811 | 0.13 |
ENST00000429586.2 ENST00000292808.5 |
ABCF3 |
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr19_+_47104553 | 0.13 |
ENST00000598871.1 ENST00000594523.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr11_-_64013288 | 0.12 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr14_-_94854926 | 0.12 |
ENST00000402629.1 ENST00000556091.1 ENST00000554720.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr12_-_49351148 | 0.12 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr1_-_113247543 | 0.11 |
ENST00000414971.1 ENST00000534717.1 |
RHOC |
ras homolog family member C |
| chr9_+_140135665 | 0.11 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr2_-_89521942 | 0.11 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chrX_+_65382381 | 0.10 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr20_+_42187608 | 0.10 |
ENST00000373100.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr17_-_79481666 | 0.10 |
ENST00000575659.1 |
ACTG1 |
actin, gamma 1 |
| chr5_+_143191726 | 0.10 |
ENST00000289448.2 |
HMHB1 |
histocompatibility (minor) HB-1 |
| chr6_-_31745037 | 0.09 |
ENST00000375688.4 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr7_+_120590803 | 0.09 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr6_-_31745085 | 0.09 |
ENST00000375686.3 ENST00000447450.1 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr8_-_110988070 | 0.08 |
ENST00000524391.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr20_+_42187682 | 0.08 |
ENST00000373092.3 ENST00000373077.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr2_+_220436917 | 0.08 |
ENST00000243786.2 |
INHA |
inhibin, alpha |
| chr12_-_51402984 | 0.08 |
ENST00000545993.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr6_+_33043703 | 0.08 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr17_+_46970134 | 0.08 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr22_-_32058166 | 0.08 |
ENST00000435900.1 ENST00000336566.4 |
PISD |
phosphatidylserine decarboxylase |
| chr9_-_114246635 | 0.08 |
ENST00000338205.5 |
KIAA0368 |
KIAA0368 |
| chrX_+_99839799 | 0.07 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr2_-_169887827 | 0.07 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr3_+_9958758 | 0.07 |
ENST00000383812.4 ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC |
interleukin 17 receptor C |
| chr17_+_46970127 | 0.07 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_+_159436120 | 0.07 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr14_+_73525229 | 0.07 |
ENST00000527432.1 ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25 |
RNA binding motif protein 25 |
| chr17_+_46970178 | 0.07 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_-_18161870 | 0.07 |
ENST00000579294.1 ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII |
flightless I homolog (Drosophila) |
| chr14_-_23588816 | 0.06 |
ENST00000206513.5 |
CEBPE |
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr17_-_79881408 | 0.06 |
ENST00000392366.3 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr2_-_238499337 | 0.06 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr22_+_23487513 | 0.06 |
ENST00000263116.2 ENST00000341989.4 |
RAB36 |
RAB36, member RAS oncogene family |
| chr6_+_2988847 | 0.06 |
ENST00000380472.3 ENST00000605901.1 ENST00000454015.1 |
NQO2 LINC01011 |
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr14_-_91710852 | 0.06 |
ENST00000535815.1 ENST00000529102.1 |
GPR68 |
G protein-coupled receptor 68 |
| chr2_-_119605253 | 0.05 |
ENST00000295206.6 |
EN1 |
engrailed homeobox 1 |
| chr12_-_49351228 | 0.05 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr18_+_268148 | 0.05 |
ENST00000581677.1 |
RP11-705O1.8 |
RP11-705O1.8 |
| chr8_-_30585217 | 0.05 |
ENST00000520888.1 ENST00000414019.1 |
GSR |
glutathione reductase |
| chr5_+_81575281 | 0.05 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 25.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 4.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 27.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 4.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.8 | 3.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 5.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 1.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 3.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 4.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 0.9 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 1.0 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.2 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 8.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0050544 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 1.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 25.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.0 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.8 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.9 | 4.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 3.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.9 | 25.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 2.4 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.6 | 3.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 2.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 1.2 | GO:2000523 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.4 | 1.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 1.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 0.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 0.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 0.9 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 4.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.3 | 1.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 0.7 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 2.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.7 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 1.9 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 2.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 5.0 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.9 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.2 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 2.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 1.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 2.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.9 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 3.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 2.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0036035 | osteoclast development(GO:0036035) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 20.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 7.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 0.9 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.2 | 2.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.8 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.0 | 1.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |