Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
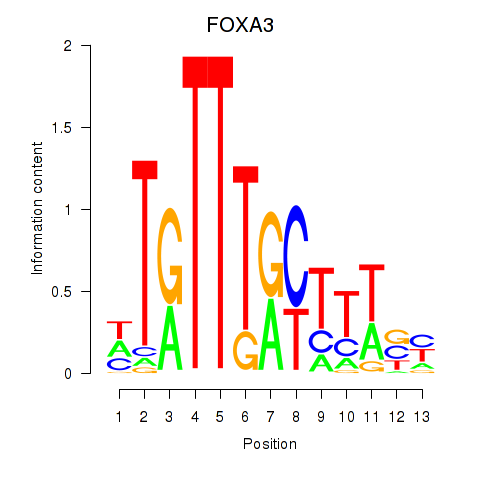
Results for FOXA3_FOXC2
Z-value: 0.76
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | FOXA3 |
|
FOXC2
|
ENSG00000176692.4 | FOXC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | -0.44 | 8.9e-02 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_116708302 | 4.42 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr17_+_79953310 | 4.23 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr17_-_26694979 | 3.71 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr17_-_26695013 | 3.68 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr6_-_41715128 | 3.62 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr1_-_169555779 | 2.91 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr17_+_72426891 | 2.87 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_128175997 | 2.50 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr2_-_21266935 | 2.44 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr8_+_97597148 | 2.35 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr14_+_21156915 | 2.02 |
ENST00000397990.4 ENST00000555597.1 |
ANG RNASE4 |
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr22_+_21128167 | 1.93 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr4_+_74269956 | 1.88 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr2_+_58655461 | 1.70 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr14_-_94789663 | 1.68 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr1_-_146696901 | 1.31 |
ENST00000369272.3 ENST00000441068.2 |
FMO5 |
flavin containing monooxygenase 5 |
| chr14_-_25479811 | 1.24 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr18_-_25616519 | 1.13 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr17_-_39684550 | 1.04 |
ENST00000455635.1 ENST00000361566.3 |
KRT19 |
keratin 19 |
| chr4_-_70080449 | 0.99 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chrX_+_9431324 | 0.97 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr7_+_134832808 | 0.94 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr17_+_72427477 | 0.93 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_57431679 | 0.79 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr11_-_115375107 | 0.78 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chrX_-_20236970 | 0.75 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr4_+_187187098 | 0.73 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr14_-_38064198 | 0.69 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr7_-_107643674 | 0.68 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr1_+_207277590 | 0.67 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr9_-_75567962 | 0.66 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr7_-_22259845 | 0.65 |
ENST00000420196.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr16_-_87970122 | 0.64 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr4_+_79567314 | 0.63 |
ENST00000503539.1 ENST00000504675.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr18_-_52626622 | 0.63 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr17_-_26697304 | 0.61 |
ENST00000536498.1 |
VTN |
vitronectin |
| chr2_+_220042933 | 0.57 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chr20_+_44519948 | 0.57 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chr4_+_79567362 | 0.57 |
ENST00000512322.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr20_+_44520009 | 0.55 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr21_-_43786634 | 0.54 |
ENST00000291527.2 |
TFF1 |
trefoil factor 1 |
| chr20_+_45338126 | 0.53 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr14_+_74034310 | 0.53 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr4_-_186696425 | 0.51 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_+_10528560 | 0.50 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr3_-_99833333 | 0.47 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr16_-_15474904 | 0.47 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr16_+_14802801 | 0.47 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr17_+_4613776 | 0.44 |
ENST00000269260.2 |
ARRB2 |
arrestin, beta 2 |
| chr5_+_156712372 | 0.44 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_73771844 | 0.42 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr19_+_782755 | 0.41 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr17_+_4613918 | 0.40 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chrX_+_23352133 | 0.40 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr10_-_63995871 | 0.39 |
ENST00000315289.2 |
RTKN2 |
rhotekin 2 |
| chr20_+_11898507 | 0.37 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr7_+_112063192 | 0.36 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr12_+_120740119 | 0.35 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr17_+_74372662 | 0.33 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr19_+_15160130 | 0.31 |
ENST00000427043.3 |
CASP14 |
caspase 14, apoptosis-related cysteine peptidase |
| chr14_-_74551096 | 0.31 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr2_-_165424973 | 0.31 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr1_-_151431909 | 0.31 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr20_-_43150601 | 0.31 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr1_+_114471972 | 0.31 |
ENST00000369559.4 ENST00000369554.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr14_-_74551172 | 0.30 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr3_-_114343039 | 0.30 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr20_+_44035200 | 0.28 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_-_46615498 | 0.28 |
ENST00000533727.1 ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
| chr1_+_114471809 | 0.27 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr4_-_140223670 | 0.27 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr7_+_73242490 | 0.27 |
ENST00000431918.1 |
CLDN4 |
claudin 4 |
| chr1_-_151431647 | 0.26 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr14_+_23299088 | 0.25 |
ENST00000355151.5 ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52 |
mitochondrial ribosomal protein L52 |
| chr2_-_175260368 | 0.25 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chrX_+_107288197 | 0.25 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr6_+_158733692 | 0.24 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr3_-_105588231 | 0.23 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_+_113885138 | 0.23 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr17_-_202579 | 0.23 |
ENST00000577079.1 ENST00000331302.7 ENST00000536489.2 |
RPH3AL |
rabphilin 3A-like (without C2 domains) |
| chr4_-_140223614 | 0.22 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr7_+_138943265 | 0.21 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr5_-_115872142 | 0.21 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_73848086 | 0.21 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr12_+_58087901 | 0.21 |
ENST00000315970.7 ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9 |
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr20_+_44035847 | 0.21 |
ENST00000372712.2 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr5_-_98262240 | 0.21 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chrX_+_107288239 | 0.20 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr6_-_122792919 | 0.19 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr2_-_65593784 | 0.19 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr8_-_28243934 | 0.19 |
ENST00000521185.1 ENST00000520290.1 ENST00000344423.5 |
ZNF395 |
zinc finger protein 395 |
| chr7_+_134576317 | 0.19 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr16_+_77233294 | 0.18 |
ENST00000378644.4 |
SYCE1L |
synaptonemal complex central element protein 1-like |
| chr3_+_142315225 | 0.17 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr7_+_28452130 | 0.17 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr10_-_46030841 | 0.17 |
ENST00000453424.2 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr12_-_71551652 | 0.17 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr1_+_29563011 | 0.16 |
ENST00000345512.3 ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU |
protein tyrosine phosphatase, receptor type, U |
| chr1_-_150669604 | 0.16 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr14_-_31926701 | 0.16 |
ENST00000310850.4 |
DTD2 |
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr12_+_59989791 | 0.16 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr16_-_86542455 | 0.16 |
ENST00000595886.1 ENST00000597578.1 ENST00000593604.1 |
FENDRR |
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr19_-_46088068 | 0.15 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr19_+_47421933 | 0.15 |
ENST00000404338.3 |
ARHGAP35 |
Rho GTPase activating protein 35 |
| chr7_-_121784285 | 0.15 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr7_-_25268104 | 0.15 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr4_+_184365744 | 0.15 |
ENST00000504169.1 ENST00000302350.4 |
CDKN2AIP |
CDKN2A interacting protein |
| chr1_-_152332480 | 0.15 |
ENST00000388718.5 |
FLG2 |
filaggrin family member 2 |
| chr1_+_154540246 | 0.14 |
ENST00000368476.3 |
CHRNB2 |
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr14_+_20811722 | 0.14 |
ENST00000429687.3 |
PARP2 |
poly (ADP-ribose) polymerase 2 |
| chr16_+_20775358 | 0.13 |
ENST00000440284.2 |
ACSM3 |
acyl-CoA synthetase medium-chain family member 3 |
| chr6_-_25830785 | 0.13 |
ENST00000468082.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chr1_-_223308098 | 0.13 |
ENST00000342210.6 |
TLR5 |
toll-like receptor 5 |
| chr7_+_134430212 | 0.13 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr12_+_72080253 | 0.13 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chr14_+_20811766 | 0.12 |
ENST00000250416.5 ENST00000527915.1 |
PARP2 |
poly (ADP-ribose) polymerase 2 |
| chr7_-_107443652 | 0.12 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr4_+_118955500 | 0.12 |
ENST00000296499.5 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr7_+_95115210 | 0.12 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr6_-_13621126 | 0.12 |
ENST00000600057.1 |
AL441883.1 |
Uncharacterized protein |
| chr3_-_186080012 | 0.12 |
ENST00000544847.1 ENST00000265022.3 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
| chr10_+_123923105 | 0.12 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr8_+_67579807 | 0.12 |
ENST00000519289.1 ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3 C8orf44 |
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr19_-_15443318 | 0.11 |
ENST00000360016.5 |
BRD4 |
bromodomain containing 4 |
| chr11_-_82708519 | 0.11 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr6_-_26056695 | 0.11 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chrX_+_65382381 | 0.10 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr2_+_136343820 | 0.10 |
ENST00000410054.1 |
R3HDM1 |
R3H domain containing 1 |
| chr6_+_108882069 | 0.10 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr1_-_28384598 | 0.10 |
ENST00000373864.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr2_-_183291741 | 0.10 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_44355257 | 0.10 |
ENST00000371477.3 |
CDC5L |
cell division cycle 5-like |
| chrX_+_24167746 | 0.10 |
ENST00000428571.1 ENST00000539115.1 |
ZFX |
zinc finger protein, X-linked |
| chr3_+_171561127 | 0.10 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr5_-_58571935 | 0.09 |
ENST00000503258.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr8_-_120651020 | 0.09 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_+_28974603 | 0.09 |
ENST00000441461.1 ENST00000358506.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr16_+_69958887 | 0.09 |
ENST00000568684.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_88720698 | 0.09 |
ENST00000226284.5 |
IBSP |
integrin-binding sialoprotein |
| chr7_+_106505696 | 0.09 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr15_-_66797172 | 0.09 |
ENST00000569438.1 ENST00000569696.1 ENST00000307961.6 |
RPL4 |
ribosomal protein L4 |
| chr1_+_87797351 | 0.09 |
ENST00000370542.1 |
LMO4 |
LIM domain only 4 |
| chr3_+_141106643 | 0.09 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr8_-_80993010 | 0.08 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr1_+_150488205 | 0.08 |
ENST00000416894.1 |
LINC00568 |
long intergenic non-protein coding RNA 568 |
| chr6_-_49755019 | 0.08 |
ENST00000304801.3 |
PGK2 |
phosphoglycerate kinase 2 |
| chr16_+_67381263 | 0.08 |
ENST00000541146.1 ENST00000563189.1 ENST00000290940.7 |
LRRC36 |
leucine rich repeat containing 36 |
| chr1_+_18958008 | 0.08 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr8_+_19171128 | 0.08 |
ENST00000265807.3 |
SH2D4A |
SH2 domain containing 4A |
| chr5_-_24645078 | 0.08 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr4_-_116034979 | 0.08 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr14_+_37126765 | 0.08 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr6_+_161123270 | 0.08 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr2_-_86422523 | 0.07 |
ENST00000442664.2 ENST00000409051.2 ENST00000449247.2 |
IMMT |
inner membrane protein, mitochondrial |
| chrX_+_24167828 | 0.07 |
ENST00000379188.3 ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX |
zinc finger protein, X-linked |
| chr1_+_18957500 | 0.07 |
ENST00000375375.3 |
PAX7 |
paired box 7 |
| chrX_+_99839799 | 0.07 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr3_+_148447887 | 0.07 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr3_-_113897899 | 0.07 |
ENST00000383673.2 ENST00000295881.7 |
DRD3 |
dopamine receptor D3 |
| chr11_-_82708435 | 0.06 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr3_+_137728842 | 0.06 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr3_+_35721106 | 0.06 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr9_-_95186739 | 0.06 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr5_+_74807886 | 0.06 |
ENST00000514296.1 |
POLK |
polymerase (DNA directed) kappa |
| chr5_+_161494770 | 0.06 |
ENST00000414552.2 ENST00000361925.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr19_+_41257084 | 0.06 |
ENST00000601393.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_162272605 | 0.06 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr21_-_36259445 | 0.06 |
ENST00000399240.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr7_+_134576151 | 0.06 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr13_-_36429763 | 0.05 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr18_-_31803169 | 0.05 |
ENST00000590712.1 |
NOL4 |
nucleolar protein 4 |
| chr6_-_24666819 | 0.05 |
ENST00000341060.3 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
| chr11_-_72070206 | 0.05 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr17_-_36358166 | 0.05 |
ENST00000537432.1 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr6_+_52051171 | 0.05 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr3_+_148545586 | 0.05 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr7_+_106505912 | 0.05 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr9_+_131062367 | 0.04 |
ENST00000601297.1 |
AL359091.2 |
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr12_-_102591604 | 0.04 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr2_+_28974668 | 0.04 |
ENST00000296122.6 ENST00000395366.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_+_174933899 | 0.04 |
ENST00000367688.3 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr16_+_67381289 | 0.04 |
ENST00000435835.3 |
LRRC36 |
leucine rich repeat containing 36 |
| chr14_+_56127989 | 0.04 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr14_-_24664540 | 0.04 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr8_-_95449155 | 0.03 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr5_-_59481406 | 0.03 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr15_-_40600026 | 0.03 |
ENST00000456256.2 ENST00000557821.1 |
PLCB2 |
phospholipase C, beta 2 |
| chr5_+_161495038 | 0.03 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr14_-_36990354 | 0.03 |
ENST00000518149.1 |
NKX2-1 |
NK2 homeobox 1 |
| chr4_-_174256276 | 0.03 |
ENST00000296503.5 |
HMGB2 |
high mobility group box 2 |
| chr13_+_52598827 | 0.03 |
ENST00000521776.2 |
UTP14C |
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr8_+_75736761 | 0.03 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr6_+_72922505 | 0.02 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr5_+_140787600 | 0.02 |
ENST00000520790.1 |
PCDHGB6 |
protocadherin gamma subfamily B, 6 |
| chrX_-_55020511 | 0.02 |
ENST00000375006.3 ENST00000374992.2 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr6_+_72922590 | 0.02 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr7_-_81399329 | 0.02 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_87804815 | 0.02 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr12_-_53207842 | 0.02 |
ENST00000458244.2 |
KRT4 |
keratin 4 |
| chr18_+_61575200 | 0.02 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.6 | 2.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 2.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 0.8 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.3 | 4.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 3.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.1 | 4.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 2.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 2.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 6.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 4.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0044218 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 6.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 4.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.8 | 2.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.6 | 1.9 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 4.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.6 | 3.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 2.9 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.5 | 4.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 2.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 2.4 | GO:0034371 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) chylomicron remodeling(GO:0034371) |
| 0.2 | 1.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.7 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 0.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 0.8 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 0.6 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 1.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 1.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 4.6 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.9 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |