Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
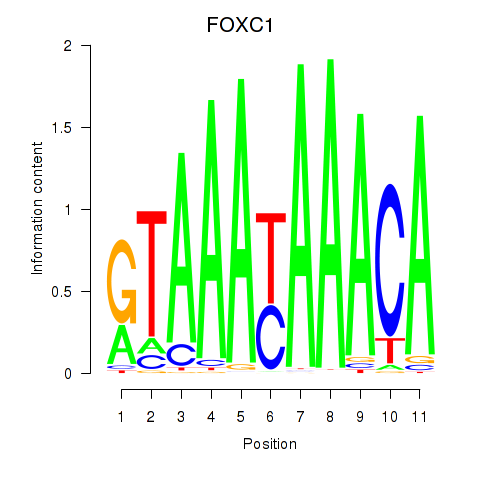
Results for FOXC1
Z-value: 1.11
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | FOXC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | 0.78 | 3.6e-04 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_54074033 | 3.37 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_-_23261589 | 3.31 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr5_-_39425222 | 3.26 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425290 | 3.17 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425068 | 2.54 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_+_71561649 | 2.49 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr2_-_190044480 | 2.45 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_-_102267953 | 2.27 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_-_93107443 | 2.15 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_-_99578789 | 2.06 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr3_-_127455200 | 1.87 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chr10_+_71561704 | 1.80 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr4_-_99578776 | 1.70 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr2_+_33359646 | 1.64 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr8_+_70404996 | 1.63 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr2_+_33359687 | 1.61 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr18_-_53303123 | 1.57 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr4_-_102268484 | 1.55 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_94079648 | 1.48 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr15_-_30114622 | 1.44 |
ENST00000495972.2 ENST00000346128.6 |
TJP1 |
tight junction protein 1 |
| chr3_-_168865522 | 1.37 |
ENST00000464456.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr4_-_102268628 | 1.36 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr4_+_86396265 | 1.33 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_-_208030647 | 1.29 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr10_+_71561630 | 1.27 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr10_+_114709999 | 1.23 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_-_95064264 | 1.22 |
ENST00000536183.1 ENST00000433091.2 ENST00000222572.3 |
PON2 |
paraoxonase 2 |
| chr15_+_96869165 | 1.19 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_16524357 | 1.18 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr17_-_46688334 | 1.13 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr8_+_31496809 | 1.11 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr8_+_31497271 | 1.06 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr1_-_100231349 | 1.04 |
ENST00000287474.5 ENST00000414213.1 |
FRRS1 |
ferric-chelate reductase 1 |
| chr1_-_94312706 | 0.96 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr7_-_27219849 | 0.95 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr8_+_54764346 | 0.95 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr12_-_49333446 | 0.94 |
ENST00000537495.1 |
AC073610.5 |
Uncharacterized protein |
| chr12_-_10251603 | 0.91 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr18_-_53070913 | 0.91 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr19_-_36523709 | 0.90 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr12_+_10365404 | 0.89 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr21_+_39628852 | 0.89 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_-_10251539 | 0.88 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr12_+_16109519 | 0.87 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr9_-_89562104 | 0.87 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr12_-_123756781 | 0.85 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr8_-_134309823 | 0.82 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr5_-_111091948 | 0.81 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr14_+_23340822 | 0.75 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr18_+_19749386 | 0.72 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr3_-_133969673 | 0.71 |
ENST00000427044.2 |
RYK |
receptor-like tyrosine kinase |
| chr8_-_134309335 | 0.69 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr3_-_114343039 | 0.69 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr21_+_39628655 | 0.67 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr8_-_93107696 | 0.65 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_42016385 | 0.64 |
ENST00000502771.1 ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3 |
cyclin D3 |
| chr15_-_72563585 | 0.61 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr15_-_55657428 | 0.60 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr1_-_169703203 | 0.59 |
ENST00000333360.7 ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE |
selectin E |
| chr8_+_120079478 | 0.57 |
ENST00000332843.2 |
COLEC10 |
collectin sub-family member 10 (C-type lectin) |
| chr6_-_31651817 | 0.55 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr2_+_210517895 | 0.55 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr3_-_114477787 | 0.51 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_-_114477962 | 0.51 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_-_57045228 | 0.51 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr10_-_118032697 | 0.50 |
ENST00000439649.3 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr22_-_38699003 | 0.50 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chr6_-_152489484 | 0.50 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr11_-_6440283 | 0.50 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_-_15382875 | 0.50 |
ENST00000408919.3 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr11_-_6440624 | 0.49 |
ENST00000311051.3 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr12_+_27175476 | 0.48 |
ENST00000546323.1 ENST00000282892.3 |
MED21 |
mediator complex subunit 21 |
| chr1_+_220701456 | 0.46 |
ENST00000366918.4 ENST00000402574.1 |
MARK1 |
MAP/microtubule affinity-regulating kinase 1 |
| chr7_-_42951509 | 0.44 |
ENST00000438029.1 ENST00000432637.1 ENST00000447342.1 ENST00000431882.2 ENST00000350427.4 ENST00000425683.1 |
C7orf25 |
chromosome 7 open reading frame 25 |
| chr10_-_118031778 | 0.41 |
ENST00000369236.1 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr4_+_71587669 | 0.39 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr16_-_53737795 | 0.39 |
ENST00000262135.4 ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L |
RPGRIP1-like |
| chr15_+_67418047 | 0.39 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr16_-_53737722 | 0.37 |
ENST00000569716.1 ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L |
RPGRIP1-like |
| chr3_-_49131788 | 0.37 |
ENST00000395443.2 ENST00000411682.1 |
QRICH1 |
glutamine-rich 1 |
| chr10_-_118032979 | 0.37 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr12_+_122516626 | 0.37 |
ENST00000319080.7 |
MLXIP |
MLX interacting protein |
| chr2_-_71454185 | 0.35 |
ENST00000244221.8 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
| chr4_-_186696425 | 0.34 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr7_-_14028488 | 0.34 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr6_-_152639479 | 0.34 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr3_+_46448648 | 0.33 |
ENST00000399036.3 |
CCRL2 |
chemokine (C-C motif) receptor-like 2 |
| chr7_-_14029283 | 0.32 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr7_-_72972319 | 0.32 |
ENST00000223368.2 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr19_-_4831701 | 0.31 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr10_+_71562180 | 0.31 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr1_+_43855560 | 0.31 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr11_+_27076764 | 0.31 |
ENST00000525090.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr16_+_8715536 | 0.31 |
ENST00000563958.1 ENST00000381920.3 ENST00000564554.1 |
METTL22 |
methyltransferase like 22 |
| chr6_+_101846664 | 0.30 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr16_+_8715574 | 0.29 |
ENST00000561758.1 |
METTL22 |
methyltransferase like 22 |
| chr11_-_66112555 | 0.29 |
ENST00000425825.2 ENST00000359957.3 |
BRMS1 |
breast cancer metastasis suppressor 1 |
| chr8_-_93107827 | 0.28 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr16_+_56969284 | 0.28 |
ENST00000568358.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr16_+_6069072 | 0.28 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_+_6069586 | 0.27 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_189349162 | 0.27 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr15_-_50411412 | 0.27 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr1_+_25664408 | 0.26 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr1_+_155099927 | 0.26 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr2_+_207024306 | 0.25 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_161056762 | 0.25 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr4_-_164534657 | 0.25 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr3_+_46449049 | 0.25 |
ENST00000357392.4 ENST00000400880.3 ENST00000433848.1 |
CCRL2 |
chemokine (C-C motif) receptor-like 2 |
| chrX_+_54947229 | 0.25 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr2_+_173686303 | 0.24 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_44502597 | 0.24 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr7_+_79998864 | 0.23 |
ENST00000435819.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr10_+_13652047 | 0.23 |
ENST00000601460.1 |
RP11-295P9.3 |
Uncharacterized protein |
| chrX_+_38211777 | 0.23 |
ENST00000039007.4 |
OTC |
ornithine carbamoyltransferase |
| chr10_+_95517660 | 0.22 |
ENST00000371413.3 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr3_+_178866199 | 0.22 |
ENST00000263967.3 |
PIK3CA |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr2_-_161056802 | 0.22 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr17_+_57642886 | 0.21 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr8_-_19540086 | 0.21 |
ENST00000332246.6 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_-_83765613 | 0.21 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr13_+_27825706 | 0.21 |
ENST00000272274.4 ENST00000319826.4 ENST00000326092.4 |
RPL21 |
ribosomal protein L21 |
| chr3_-_48481434 | 0.21 |
ENST00000395694.2 ENST00000447018.1 ENST00000442740.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr3_-_128902729 | 0.21 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr3_+_48481658 | 0.20 |
ENST00000438607.2 |
TMA7 |
translation machinery associated 7 homolog (S. cerevisiae) |
| chr3_+_171758344 | 0.20 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr16_-_4817129 | 0.19 |
ENST00000545009.1 ENST00000219478.6 |
ZNF500 |
zinc finger protein 500 |
| chr4_-_74486347 | 0.19 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_-_246729544 | 0.18 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr1_+_73771844 | 0.18 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr13_+_27825446 | 0.18 |
ENST00000311549.6 |
RPL21 |
ribosomal protein L21 |
| chr15_+_58430567 | 0.18 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr3_-_48481518 | 0.17 |
ENST00000412398.2 ENST00000395696.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr22_-_43036607 | 0.17 |
ENST00000505920.1 |
ATP5L2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr1_+_18958008 | 0.16 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr2_+_181845843 | 0.16 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr8_-_141931287 | 0.16 |
ENST00000517887.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr5_+_179921344 | 0.15 |
ENST00000261951.4 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr19_-_52255107 | 0.15 |
ENST00000595042.1 ENST00000304748.4 |
FPR1 |
formyl peptide receptor 1 |
| chr17_-_19290483 | 0.15 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr15_+_80351977 | 0.15 |
ENST00000559157.1 ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr7_+_80231466 | 0.15 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr6_-_52926539 | 0.14 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr5_+_179921430 | 0.13 |
ENST00000393356.1 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr20_-_35890211 | 0.12 |
ENST00000373614.2 |
GHRH |
growth hormone releasing hormone |
| chr19_+_36024310 | 0.11 |
ENST00000222286.4 |
GAPDHS |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr20_-_17511962 | 0.11 |
ENST00000377873.3 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr12_+_54378923 | 0.11 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr2_-_152146385 | 0.10 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr12_-_92539614 | 0.10 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr2_-_224467093 | 0.10 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chrX_-_138790348 | 0.10 |
ENST00000414978.1 ENST00000519895.1 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr14_-_92572894 | 0.09 |
ENST00000532032.1 ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3 |
ataxin 3 |
| chr1_-_43855444 | 0.09 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr5_-_146302078 | 0.09 |
ENST00000508545.2 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_27886676 | 0.09 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr9_+_71944241 | 0.09 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr12_+_99038919 | 0.08 |
ENST00000551964.1 |
APAF1 |
apoptotic peptidase activating factor 1 |
| chr11_-_790060 | 0.08 |
ENST00000330106.4 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
| chr13_+_52598827 | 0.07 |
ENST00000521776.2 |
UTP14C |
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr22_+_41487711 | 0.07 |
ENST00000263253.7 |
EP300 |
E1A binding protein p300 |
| chr15_+_58430368 | 0.06 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr1_-_159684371 | 0.06 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr1_-_19811132 | 0.06 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr20_+_36946029 | 0.06 |
ENST00000417318.1 |
BPI |
bactericidal/permeability-increasing protein |
| chr10_+_95517616 | 0.05 |
ENST00000371418.4 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr17_+_7533439 | 0.04 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr1_-_149982624 | 0.04 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
| chr6_+_45296391 | 0.03 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr13_-_36050819 | 0.03 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr20_+_44509857 | 0.03 |
ENST00000372523.1 ENST00000372520.1 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
| chr11_-_108464321 | 0.03 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr1_-_116383738 | 0.02 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr9_-_5185629 | 0.02 |
ENST00000381641.3 |
INSL6 |
insulin-like 6 |
| chr15_+_80351910 | 0.02 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr10_+_95517566 | 0.02 |
ENST00000542308.1 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr18_+_21032781 | 0.01 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr3_-_11623804 | 0.01 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr11_+_8040739 | 0.01 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr1_-_150669500 | 0.01 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr11_-_108464465 | 0.00 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr2_-_8977714 | 0.00 |
ENST00000319688.5 ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220 |
kinase D-interacting substrate, 220kDa |
| chr11_+_112832090 | 0.00 |
ENST00000533760.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr3_-_195310802 | 0.00 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 3.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 5.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 9.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.3 | 3.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 0.9 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 0.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 2.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.6 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.0 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 5.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.0 | 9.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.8 | 2.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 5.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 0.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 3.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.0 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 3.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 2.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 9.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 8.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 9.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.7 | 5.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.5 | 5.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.5 | 1.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 3.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 2.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 1.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.3 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 2.9 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.7 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.2 | 1.2 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 1.5 | GO:0090232 | peripheral nervous system myelin maintenance(GO:0032287) positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 3.7 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 5.8 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.6 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.4 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.1 | 0.3 | GO:0003180 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.1 | 0.8 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 3.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.2 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 1.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.4 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.9 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.3 | GO:0045350 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) positive regulation of chemokine biosynthetic process(GO:0045080) interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 1.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.7 | GO:0048934 | mechanosensory behavior(GO:0007638) peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |