Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
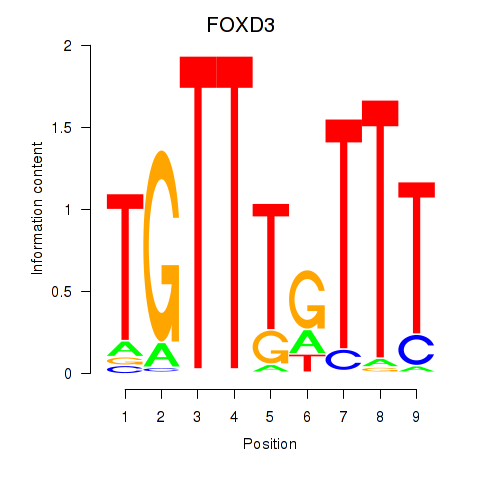
Results for FOXD3_FOXI1_FOXF1
Z-value: 1.33
Transcription factors associated with FOXD3_FOXI1_FOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD3
|
ENSG00000187140.4 | FOXD3 |
|
FOXI1
|
ENSG00000168269.7 | FOXI1 |
|
FOXF1
|
ENSG00000103241.5 | FOXF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXF1 | hg19_v2_chr16_+_86544113_86544145 | 0.29 | 2.7e-01 | Click! |
| FOXD3 | hg19_v2_chr1_+_63788730_63788730 | -0.22 | 4.2e-01 | Click! |
| FOXI1 | hg19_v2_chr5_+_169532896_169532917 | 0.03 | 9.1e-01 | Click! |
Activity profile of FOXD3_FOXI1_FOXF1 motif
Sorted Z-values of FOXD3_FOXI1_FOXF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD3_FOXI1_FOXF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111091948 | 3.14 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr3_-_16524357 | 3.04 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr18_-_53070913 | 2.76 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr13_-_47012325 | 2.57 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr4_-_84035868 | 2.48 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr4_+_68424434 | 2.38 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr2_-_152146385 | 2.37 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr4_-_84035905 | 2.33 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr5_-_88179302 | 2.32 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_-_158345462 | 2.09 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr17_-_29641104 | 1.76 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr1_+_150122034 | 1.75 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr4_-_105416039 | 1.66 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr17_-_29641084 | 1.65 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr10_-_98031265 | 1.57 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr6_-_133084580 | 1.53 |
ENST00000525270.1 ENST00000530536.1 ENST00000524919.1 |
VNN2 |
vanin 2 |
| chr10_-_98031310 | 1.53 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr12_+_59989791 | 1.42 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr20_+_9494987 | 1.40 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr3_-_121379739 | 1.38 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr1_-_173174681 | 1.33 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr8_-_108510224 | 1.31 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr15_+_89181974 | 1.30 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 1.27 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr1_-_27952741 | 1.25 |
ENST00000399173.1 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr15_+_89182178 | 1.24 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr5_-_54281407 | 1.24 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr1_+_111415757 | 1.23 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr7_+_119913688 | 1.21 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr10_-_14050522 | 1.20 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr8_+_79428539 | 1.20 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr12_-_15038779 | 1.19 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr17_-_46688334 | 1.16 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr18_-_53253323 | 1.13 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr6_+_114178512 | 1.13 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr8_-_120685608 | 1.11 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_+_127413677 | 1.10 |
ENST00000356887.7 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr18_-_53253112 | 1.10 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr7_-_144435985 | 1.07 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr12_-_49333446 | 1.04 |
ENST00000537495.1 |
AC073610.5 |
Uncharacterized protein |
| chr1_+_174844645 | 1.03 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr3_-_127455200 | 1.00 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chrX_+_9431324 | 0.99 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr3_-_119278376 | 0.98 |
ENST00000478182.1 |
CD80 |
CD80 molecule |
| chr18_-_53089723 | 0.98 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr10_-_90712520 | 0.97 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr10_+_35484793 | 0.96 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr12_-_92539614 | 0.96 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr1_+_32739733 | 0.96 |
ENST00000333070.4 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr2_+_33661382 | 0.95 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr6_+_89790490 | 0.94 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr1_+_192544857 | 0.93 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chrX_+_24483338 | 0.92 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chrX_-_70331298 | 0.91 |
ENST00000456850.2 ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG |
interleukin 2 receptor, gamma |
| chr18_-_53177984 | 0.90 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr7_-_37026108 | 0.88 |
ENST00000396045.3 |
ELMO1 |
engulfment and cell motility 1 |
| chr3_+_119316689 | 0.88 |
ENST00000273371.4 |
PLA1A |
phospholipase A1 member A |
| chr6_+_31582961 | 0.87 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
| chr6_-_131291572 | 0.87 |
ENST00000529208.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr8_-_120651020 | 0.86 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr21_-_15918618 | 0.85 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr3_-_119278446 | 0.85 |
ENST00000264246.3 |
CD80 |
CD80 molecule |
| chr2_-_175711133 | 0.84 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_-_144533074 | 0.84 |
ENST00000360057.3 ENST00000378099.3 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr16_-_24942655 | 0.84 |
ENST00000573765.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr14_-_25479811 | 0.83 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr5_-_111093167 | 0.83 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr13_-_46756351 | 0.82 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_-_152639479 | 0.81 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_19811132 | 0.81 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr1_-_160681593 | 0.80 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chrX_-_154563889 | 0.77 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr6_-_26056695 | 0.77 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr13_-_46716969 | 0.77 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr16_-_24942273 | 0.77 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr1_-_57045228 | 0.76 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr17_+_67498538 | 0.76 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr15_-_55562582 | 0.76 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr11_-_73693875 | 0.75 |
ENST00000536983.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr6_-_46922659 | 0.75 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chr3_-_119276575 | 0.74 |
ENST00000383669.3 ENST00000383668.3 |
CD80 |
CD80 molecule |
| chr15_-_37390482 | 0.74 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr13_-_99959641 | 0.73 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr3_+_119316721 | 0.73 |
ENST00000488919.1 ENST00000495992.1 |
PLA1A |
phospholipase A1 member A |
| chrX_-_70329118 | 0.72 |
ENST00000374188.3 |
IL2RG |
interleukin 2 receptor, gamma |
| chr3_+_178276488 | 0.72 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr6_+_12958137 | 0.72 |
ENST00000457702.2 ENST00000379345.2 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr1_-_150669500 | 0.72 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr11_-_78052923 | 0.72 |
ENST00000340149.2 |
GAB2 |
GRB2-associated binding protein 2 |
| chr5_+_156712372 | 0.72 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr3_-_114035026 | 0.71 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr13_+_50070077 | 0.71 |
ENST00000378319.3 ENST00000426879.1 |
PHF11 |
PHD finger protein 11 |
| chr18_+_77160282 | 0.70 |
ENST00000318065.5 ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr14_+_61654271 | 0.69 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr10_+_31610064 | 0.69 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr12_+_48513009 | 0.67 |
ENST00000359794.5 ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM |
phosphofructokinase, muscle |
| chr14_-_23285011 | 0.66 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_+_116654935 | 0.66 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr11_-_93276582 | 0.65 |
ENST00000298966.2 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
| chr4_-_38806404 | 0.65 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr1_-_92371839 | 0.64 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr11_+_93479588 | 0.64 |
ENST00000526335.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
| chr6_-_152489484 | 0.63 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr12_-_91576561 | 0.63 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr11_-_46142948 | 0.63 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr17_-_19290483 | 0.62 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr12_-_9913489 | 0.62 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr3_-_127541194 | 0.62 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr6_+_32812568 | 0.61 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr22_-_17680472 | 0.61 |
ENST00000330232.4 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
| chr7_-_86849025 | 0.61 |
ENST00000257637.3 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr6_+_89790459 | 0.61 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr18_-_53069419 | 0.61 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr3_-_114343039 | 0.60 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_-_94417339 | 0.60 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr11_+_65657875 | 0.60 |
ENST00000312579.2 |
CCDC85B |
coiled-coil domain containing 85B |
| chr13_-_99910673 | 0.60 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr7_-_37488834 | 0.59 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr14_-_23285069 | 0.59 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_92414055 | 0.59 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr4_-_103266626 | 0.59 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr14_-_23288930 | 0.58 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr18_-_53303123 | 0.58 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr2_+_152214098 | 0.57 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr12_+_59989918 | 0.57 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_+_10365404 | 0.56 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr2_+_58655461 | 0.56 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr8_+_70404996 | 0.55 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr10_-_46030841 | 0.55 |
ENST00000453424.2 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr11_+_5710919 | 0.55 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr1_+_66797687 | 0.54 |
ENST00000371045.5 ENST00000531025.1 ENST00000526197.1 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr1_+_198608146 | 0.54 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr3_+_157154578 | 0.54 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr9_-_16727978 | 0.53 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr1_-_170043709 | 0.53 |
ENST00000367767.1 ENST00000361580.2 ENST00000538366.1 |
KIFAP3 |
kinesin-associated protein 3 |
| chr3_+_152017360 | 0.53 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr15_-_55562479 | 0.52 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_-_68696652 | 0.52 |
ENST00000539972.1 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr8_+_74903580 | 0.51 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr19_-_11689752 | 0.50 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr7_+_142985308 | 0.49 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr2_-_191878162 | 0.49 |
ENST00000540176.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr3_-_105588231 | 0.48 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr11_-_72070206 | 0.48 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr4_+_184365744 | 0.48 |
ENST00000504169.1 ENST00000302350.4 |
CDKN2AIP |
CDKN2A interacting protein |
| chr10_-_94003003 | 0.48 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr7_+_120590803 | 0.47 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr11_-_62323702 | 0.47 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr15_-_55563072 | 0.47 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr7_+_28452130 | 0.47 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr9_+_2029019 | 0.47 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_26321284 | 0.47 |
ENST00000506956.1 ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_16761678 | 0.46 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr1_-_32801825 | 0.46 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr12_-_15114492 | 0.46 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_207095324 | 0.45 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr12_-_15114603 | 0.45 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_+_68665608 | 0.45 |
ENST00000509734.1 ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17 |
RAD17 homolog (S. pombe) |
| chr5_-_142782862 | 0.44 |
ENST00000415690.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_+_201994569 | 0.44 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr18_+_32556892 | 0.44 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_164528866 | 0.43 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr4_+_17578815 | 0.43 |
ENST00000226299.4 |
LAP3 |
leucine aminopeptidase 3 |
| chr12_-_91576429 | 0.43 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr12_+_54892550 | 0.43 |
ENST00000545638.2 |
NCKAP1L |
NCK-associated protein 1-like |
| chr7_+_80231466 | 0.42 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr14_+_65878565 | 0.42 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr12_+_12878829 | 0.42 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr11_+_128563948 | 0.42 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_89378261 | 0.42 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr8_-_95449155 | 0.42 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr9_-_20622478 | 0.41 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr9_+_2015335 | 0.41 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_226411319 | 0.40 |
ENST00000542034.1 ENST00000366810.5 |
MIXL1 |
Mix paired-like homeobox |
| chr1_-_226926864 | 0.40 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr17_-_37934466 | 0.40 |
ENST00000583368.1 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
| chr4_+_146403912 | 0.40 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr3_+_141106643 | 0.39 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr20_-_5093399 | 0.39 |
ENST00000379276.4 |
TMEM230 |
transmembrane protein 230 |
| chr2_-_231989808 | 0.39 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr11_-_108093329 | 0.39 |
ENST00000278612.8 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
| chr20_-_5093673 | 0.39 |
ENST00000379299.2 ENST00000379286.2 ENST00000379279.2 ENST00000379283.2 |
TMEM230 |
transmembrane protein 230 |
| chr12_-_91574142 | 0.38 |
ENST00000547937.1 |
DCN |
decorin |
| chr6_-_112575912 | 0.37 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr20_-_5093503 | 0.37 |
ENST00000379277.2 |
TMEM230 |
transmembrane protein 230 |
| chr20_-_5093713 | 0.37 |
ENST00000342308.5 ENST00000202834.7 |
TMEM230 |
transmembrane protein 230 |
| chr13_+_76334567 | 0.37 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr3_+_69812877 | 0.37 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr21_+_30502806 | 0.37 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr13_+_76334795 | 0.37 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr11_+_63304273 | 0.36 |
ENST00000439013.2 ENST00000255688.3 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
| chr11_+_128563652 | 0.36 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_120591170 | 0.36 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr8_-_60031762 | 0.36 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr15_+_80351910 | 0.36 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr6_-_41703952 | 0.35 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr5_-_24645078 | 0.35 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr16_+_27438563 | 0.35 |
ENST00000395754.4 |
IL21R |
interleukin 21 receptor |
| chr3_-_49066811 | 0.35 |
ENST00000442157.1 ENST00000326739.4 |
IMPDH2 |
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_-_790060 | 0.35 |
ENST00000330106.4 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 8.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 8.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.6 | 2.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.5 | 1.6 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.5 | 1.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.5 | 2.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.4 | 2.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 1.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 0.8 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 1.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 0.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.5 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 1.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 2.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.8 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 2.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.4 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.7 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 1.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 5.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 3.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 3.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 2.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.5 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.8 | 2.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.6 | 2.6 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 1.9 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.4 | 1.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 3.0 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.4 | 2.6 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.3 | 1.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 2.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 1.6 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.3 | 1.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 1.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 1.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 2.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 1.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 0.8 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.3 | 4.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 0.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 1.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.1 | GO:0060938 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.2 | 0.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 0.7 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 0.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 1.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 0.5 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 | 1.4 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 1.2 | GO:0071724 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.0 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.2 | 0.8 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 0.5 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.2 | 0.5 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 0.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 1.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.4 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 1.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 2.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.9 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 2.0 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.5 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.9 | GO:0071672 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 1.8 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.5 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.1 | 1.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.5 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) bone marrow development(GO:0048539) |
| 0.1 | 0.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.3 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 0.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.1 | 0.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.2 | GO:0060769 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.1 | 0.2 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.6 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.3 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.4 | GO:0045927 | positive regulation of growth(GO:0045927) |
| 0.1 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.8 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.1 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.2 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.1 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.5 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.2 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0060947 | cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.0 | GO:1903044 | protein localization to membrane raft(GO:1903044) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 8.5 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.6 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0051315 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 2.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.0 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.3 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.2 | GO:0097296 | positive regulation of hair follicle development(GO:0051798) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.2 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.2 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0044362 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 1.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 0.8 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 5.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 1.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 2.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 3.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 2.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 6.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) endosome lumen(GO:0031904) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |