Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
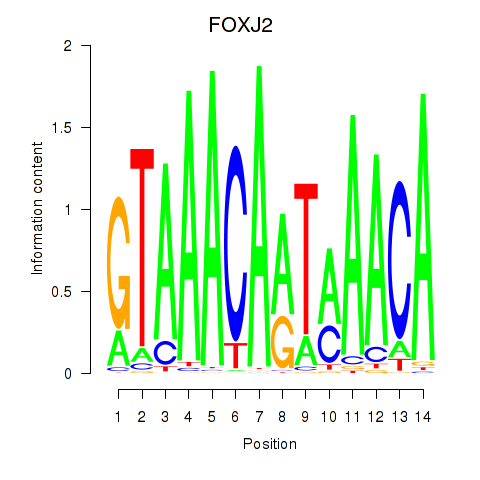
Results for FOXJ2
Z-value: 0.94
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | FOXJ2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg19_v2_chr12_+_8185288_8185339 | 0.40 | 1.2e-01 | Click! |
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_35201826 | 4.09 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr7_-_80548667 | 3.60 |
ENST00000265361.3 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_-_8815215 | 2.37 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815299 | 2.36 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr13_-_38172863 | 2.29 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr18_-_21852143 | 2.22 |
ENST00000399443.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr15_+_32933866 | 1.63 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr12_-_8815404 | 1.54 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr4_+_89299885 | 1.37 |
ENST00000380265.5 ENST00000273960.3 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr4_+_89299994 | 1.24 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_56507586 | 1.20 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr4_-_90757364 | 1.16 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_91574142 | 0.99 |
ENST00000547937.1 |
DCN |
decorin |
| chr12_-_53074182 | 0.96 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr2_-_85641162 | 0.95 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr1_-_94079648 | 0.92 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr1_+_223889285 | 0.90 |
ENST00000433674.2 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr15_+_67430339 | 0.88 |
ENST00000439724.3 |
SMAD3 |
SMAD family member 3 |
| chr17_-_46115122 | 0.84 |
ENST00000006101.4 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
| chr3_+_128444994 | 0.84 |
ENST00000482525.1 |
RAB7A |
RAB7A, member RAS oncogene family |
| chr12_+_79258444 | 0.83 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr4_-_90756769 | 0.75 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_-_95274536 | 0.69 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr12_-_8815477 | 0.68 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr8_-_80993010 | 0.66 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr16_+_3068393 | 0.62 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr3_+_128444965 | 0.62 |
ENST00000265062.3 |
RAB7A |
RAB7A, member RAS oncogene family |
| chr8_+_101170257 | 0.60 |
ENST00000251809.3 |
SPAG1 |
sperm associated antigen 1 |
| chr14_+_90863364 | 0.52 |
ENST00000447653.3 ENST00000553542.1 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr7_+_77469439 | 0.50 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr12_+_6833237 | 0.50 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr6_-_99842041 | 0.48 |
ENST00000254759.3 ENST00000369242.1 |
COQ3 |
coenzyme Q3 methyltransferase |
| chr1_-_217804377 | 0.48 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr4_-_164534657 | 0.47 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr8_-_54755789 | 0.46 |
ENST00000359530.2 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chrX_-_107682702 | 0.46 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr1_+_43855560 | 0.45 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr21_-_27423339 | 0.42 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr14_+_90863327 | 0.38 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr7_-_102283238 | 0.38 |
ENST00000340457.8 |
UPK3BL |
uroplakin 3B-like |
| chr2_+_210517895 | 0.37 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chrX_+_55744166 | 0.34 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr16_-_30440530 | 0.34 |
ENST00000568434.1 |
DCTPP1 |
dCTP pyrophosphatase 1 |
| chr3_+_157154578 | 0.33 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_-_240775447 | 0.33 |
ENST00000318160.4 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
| chr3_-_64009102 | 0.33 |
ENST00000478185.1 ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr1_+_28879588 | 0.31 |
ENST00000373830.3 |
TRNAU1AP |
tRNA selenocysteine 1 associated protein 1 |
| chr6_+_31462658 | 0.30 |
ENST00000538442.1 |
MICB |
MHC class I polypeptide-related sequence B |
| chrX_+_55744228 | 0.30 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr1_+_146373546 | 0.29 |
ENST00000446760.2 |
NBPF12 |
neuroblastoma breakpoint family, member 12 |
| chr12_-_96429423 | 0.28 |
ENST00000228740.2 |
LTA4H |
leukotriene A4 hydrolase |
| chr3_-_64009658 | 0.28 |
ENST00000394431.2 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr22_-_39268308 | 0.26 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr8_+_30244580 | 0.24 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr13_-_37573432 | 0.23 |
ENST00000413537.2 ENST00000443765.1 ENST00000239891.3 |
ALG5 |
ALG5, dolichyl-phosphate beta-glucosyltransferase |
| chr10_-_36813162 | 0.22 |
ENST00000440465.1 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
| chr19_+_13134772 | 0.20 |
ENST00000587760.1 ENST00000585575.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_38019878 | 0.20 |
ENST00000296215.6 |
SNIP1 |
Smad nuclear interacting protein 1 |
| chr12_-_57941004 | 0.19 |
ENST00000550750.1 ENST00000548249.1 |
DCTN2 |
dynactin 2 (p50) |
| chr1_+_162602244 | 0.19 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr4_+_57845024 | 0.17 |
ENST00000431623.2 ENST00000441246.2 |
POLR2B |
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr15_-_56757329 | 0.17 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr4_+_68424434 | 0.17 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr8_+_85095769 | 0.16 |
ENST00000518566.1 |
RALYL |
RALY RNA binding protein-like |
| chr1_-_43855479 | 0.16 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr12_+_21679220 | 0.16 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr12_-_68553512 | 0.16 |
ENST00000229135.3 |
IFNG |
interferon, gamma |
| chr3_-_49131788 | 0.16 |
ENST00000395443.2 ENST00000411682.1 |
QRICH1 |
glutamine-rich 1 |
| chr14_+_52456193 | 0.15 |
ENST00000261700.3 |
C14orf166 |
chromosome 14 open reading frame 166 |
| chr2_-_55237484 | 0.15 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr9_-_134615443 | 0.13 |
ENST00000372195.1 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr6_+_30539153 | 0.13 |
ENST00000326195.8 ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1 |
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr1_+_160765884 | 0.13 |
ENST00000392203.4 |
LY9 |
lymphocyte antigen 9 |
| chrX_+_107683096 | 0.13 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr1_+_160765919 | 0.12 |
ENST00000341032.4 ENST00000368041.2 ENST00000368040.1 |
LY9 |
lymphocyte antigen 9 |
| chr15_+_42697065 | 0.12 |
ENST00000565559.1 |
CAPN3 |
calpain 3, (p94) |
| chr6_-_27858570 | 0.12 |
ENST00000359303.2 |
HIST1H3J |
histone cluster 1, H3j |
| chr19_+_11658655 | 0.12 |
ENST00000588935.1 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr2_-_74619152 | 0.11 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr1_-_45452240 | 0.11 |
ENST00000372183.3 ENST00000372182.4 ENST00000360403.2 |
EIF2B3 |
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa |
| chr20_+_32782375 | 0.11 |
ENST00000568305.1 |
ASIP |
agouti signaling protein |
| chr1_+_160765947 | 0.10 |
ENST00000263285.6 ENST00000368039.2 |
LY9 |
lymphocyte antigen 9 |
| chr3_+_132036207 | 0.10 |
ENST00000336375.5 ENST00000495911.1 |
ACPP |
acid phosphatase, prostate |
| chr1_+_160765860 | 0.10 |
ENST00000368037.5 |
LY9 |
lymphocyte antigen 9 |
| chr3_+_52017454 | 0.10 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr18_+_3447572 | 0.10 |
ENST00000548489.2 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_+_52456327 | 0.09 |
ENST00000556760.1 |
C14orf166 |
chromosome 14 open reading frame 166 |
| chr2_+_231921574 | 0.09 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr15_+_66797627 | 0.09 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chrX_-_118699325 | 0.09 |
ENST00000320339.4 ENST00000371594.4 ENST00000536133.1 |
CXorf56 |
chromosome X open reading frame 56 |
| chr19_+_49866331 | 0.09 |
ENST00000597873.1 |
DKKL1 |
dickkopf-like 1 |
| chr1_-_19811132 | 0.08 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr10_+_95848824 | 0.08 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr10_-_105992059 | 0.08 |
ENST00000369720.1 ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96 |
WD repeat domain 96 |
| chr2_+_101591314 | 0.08 |
ENST00000450763.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr1_+_197237352 | 0.07 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr11_-_125550764 | 0.07 |
ENST00000527795.1 |
ACRV1 |
acrosomal vesicle protein 1 |
| chr8_+_128426535 | 0.07 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr6_-_31560729 | 0.07 |
ENST00000340027.5 ENST00000376073.4 ENST00000376072.3 |
NCR3 |
natural cytotoxicity triggering receptor 3 |
| chr1_+_145883868 | 0.07 |
ENST00000447947.2 |
GPR89C |
G protein-coupled receptor 89C |
| chr4_-_68749745 | 0.06 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr9_-_99417562 | 0.06 |
ENST00000375234.3 ENST00000446045.1 |
AAED1 |
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr19_+_47538560 | 0.06 |
ENST00000439365.2 ENST00000594670.1 |
NPAS1 |
neuronal PAS domain protein 1 |
| chr17_+_27046988 | 0.06 |
ENST00000496182.1 |
RPL23A |
ribosomal protein L23a |
| chr12_-_102874330 | 0.06 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr11_+_63137251 | 0.06 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr10_+_105005644 | 0.06 |
ENST00000441178.2 |
RP11-332O19.5 |
ribulose-5-phosphate-3-epimerase-like 1 |
| chr3_-_109056419 | 0.06 |
ENST00000335658.6 |
DPPA4 |
developmental pluripotency associated 4 |
| chr12_-_102874102 | 0.05 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr12_-_102874416 | 0.05 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr1_-_27701307 | 0.05 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr12_+_54891495 | 0.05 |
ENST00000293373.6 |
NCKAP1L |
NCK-associated protein 1-like |
| chr3_-_57233966 | 0.05 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr15_+_42697018 | 0.05 |
ENST00000397204.4 |
CAPN3 |
calpain 3, (p94) |
| chr2_+_157330081 | 0.04 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr20_+_55926583 | 0.04 |
ENST00000395840.2 |
RAE1 |
ribonucleic acid export 1 |
| chr15_-_55881135 | 0.04 |
ENST00000302000.6 |
PYGO1 |
pygopus family PHD finger 1 |
| chr1_-_230513367 | 0.04 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr4_-_38806404 | 0.04 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr2_+_90121477 | 0.04 |
ENST00000483379.1 |
IGKV1D-17 |
immunoglobulin kappa variable 1D-17 |
| chr4_-_68749699 | 0.04 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr15_+_66797455 | 0.04 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr4_+_110749143 | 0.04 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_+_123949053 | 0.03 |
ENST00000350887.5 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr14_+_37126765 | 0.03 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr2_+_216176540 | 0.03 |
ENST00000236959.9 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr18_+_32455201 | 0.03 |
ENST00000590831.2 |
DTNA |
dystrobrevin, alpha |
| chr4_-_47983519 | 0.02 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chrX_+_11311533 | 0.02 |
ENST00000380714.3 ENST00000380712.3 ENST00000348912.4 |
AMELX |
amelogenin, X-linked |
| chr16_-_90096309 | 0.02 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr8_+_58890917 | 0.02 |
ENST00000522992.1 |
RP11-1112C15.1 |
RP11-1112C15.1 |
| chr19_-_54619006 | 0.02 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr12_-_102874378 | 0.01 |
ENST00000456098.1 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr9_+_5510558 | 0.01 |
ENST00000397747.3 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr15_+_42696992 | 0.01 |
ENST00000561817.1 |
CAPN3 |
calpain 3, (p94) |
| chr1_-_220263096 | 0.01 |
ENST00000463953.1 ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1 |
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr6_+_10748019 | 0.01 |
ENST00000543878.1 ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L TMEM14B |
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr1_-_60392452 | 0.01 |
ENST00000371204.3 |
CYP2J2 |
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr6_+_152130240 | 0.01 |
ENST00000427531.2 |
ESR1 |
estrogen receptor 1 |
| chr17_+_27047244 | 0.01 |
ENST00000394938.4 ENST00000394935.3 ENST00000355731.4 |
RPL23A |
ribosomal protein L23a |
| chr6_+_10747986 | 0.00 |
ENST00000379542.5 |
TMEM14B |
transmembrane protein 14B |
| chr7_+_80275621 | 0.00 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr9_+_133569108 | 0.00 |
ENST00000372358.5 ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2 |
exosome component 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.6 | 3.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.5 | 4.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.5 | 1.5 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.4 | 1.9 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 0.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 7.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.6 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.4 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:1901668 | corpus callosum morphogenesis(GO:0021540) regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 2.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.2 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.6 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.0 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 7.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 0.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 3.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 4.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 7.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 7.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |