Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
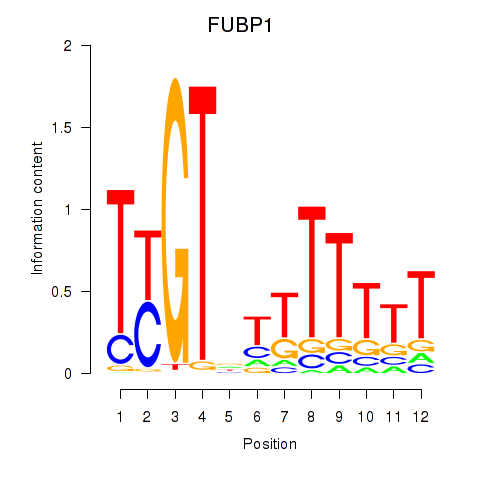
Results for FUBP1
Z-value: 0.94
Transcription factors associated with FUBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FUBP1
|
ENSG00000162613.12 | FUBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FUBP1 | hg19_v2_chr1_-_78444738_78444773 | -0.14 | 6.1e-01 | Click! |
Activity profile of FUBP1 motif
Sorted Z-values of FUBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FUBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_53255766 | 3.82 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr7_+_116166331 | 3.09 |
ENST00000393468.1 ENST00000393467.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr11_-_33891362 | 2.79 |
ENST00000395833.3 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr12_+_48513570 | 2.22 |
ENST00000551804.1 |
PFKM |
phosphofructokinase, muscle |
| chr6_+_121756809 | 2.22 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr7_-_87849340 | 2.13 |
ENST00000419179.1 ENST00000265729.2 |
SRI |
sorcin |
| chr15_+_49170083 | 1.86 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr2_+_33359646 | 1.76 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 1.75 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr4_-_16900242 | 1.49 |
ENST00000502640.1 ENST00000506732.1 |
LDB2 |
LIM domain binding 2 |
| chr4_-_25865159 | 1.36 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr4_-_16900184 | 1.27 |
ENST00000515064.1 |
LDB2 |
LIM domain binding 2 |
| chr1_+_84609944 | 1.26 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_16900217 | 1.25 |
ENST00000441778.2 |
LDB2 |
LIM domain binding 2 |
| chr4_-_16900410 | 1.21 |
ENST00000304523.5 |
LDB2 |
LIM domain binding 2 |
| chr4_-_101439242 | 1.15 |
ENST00000296420.4 |
EMCN |
endomucin |
| chr4_-_101439148 | 1.11 |
ENST00000511970.1 ENST00000502569.1 ENST00000305864.3 |
EMCN |
endomucin |
| chr14_+_61654271 | 0.94 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr7_-_124405681 | 0.92 |
ENST00000303921.2 |
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr15_-_55657428 | 0.82 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr1_+_43766642 | 0.81 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr9_+_82186872 | 0.77 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr8_+_94929969 | 0.75 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_-_120685608 | 0.73 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_-_167452298 | 0.69 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr6_+_125524785 | 0.65 |
ENST00000392482.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chrX_+_119737806 | 0.64 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chr3_-_167452262 | 0.62 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr11_+_60681346 | 0.60 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr9_-_113761720 | 0.55 |
ENST00000541779.1 ENST00000374430.2 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr7_-_144435985 | 0.55 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr9_-_110251836 | 0.55 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr12_-_91576561 | 0.55 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr7_-_83824169 | 0.55 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr21_+_30502806 | 0.54 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr3_+_173302222 | 0.51 |
ENST00000361589.4 |
NLGN1 |
neuroligin 1 |
| chr12_-_57522813 | 0.51 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr9_+_82186682 | 0.50 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chrX_+_95939638 | 0.49 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr19_+_36235964 | 0.48 |
ENST00000587708.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr12_-_91576429 | 0.48 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr7_-_144533074 | 0.46 |
ENST00000360057.3 ENST00000378099.3 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr15_+_52155001 | 0.44 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr21_-_37451680 | 0.43 |
ENST00000399201.1 |
SETD4 |
SET domain containing 4 |
| chr3_-_169587621 | 0.41 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr12_+_10365404 | 0.41 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr12_+_56324756 | 0.40 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr3_+_186383741 | 0.38 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chrX_+_52235228 | 0.37 |
ENST00000518075.1 |
XAGE1B |
X antigen family, member 1B |
| chr4_-_100242549 | 0.37 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr4_-_84205905 | 0.37 |
ENST00000311461.7 ENST00000311469.4 ENST00000439031.2 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr11_+_34073195 | 0.37 |
ENST00000341394.4 |
CAPRIN1 |
cell cycle associated protein 1 |
| chrX_+_95939711 | 0.36 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr1_-_44818599 | 0.35 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr5_-_34919094 | 0.35 |
ENST00000341754.4 |
RAD1 |
RAD1 homolog (S. pombe) |
| chr17_-_8151353 | 0.34 |
ENST00000315684.8 |
CTC1 |
CTS telomere maintenance complex component 1 |
| chr15_+_49462397 | 0.33 |
ENST00000396509.2 |
GALK2 |
galactokinase 2 |
| chr1_-_43855479 | 0.31 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr6_-_111888474 | 0.31 |
ENST00000368735.1 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr19_+_19303008 | 0.31 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr15_+_49462434 | 0.31 |
ENST00000558145.1 ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2 |
galactokinase 2 |
| chr3_-_112127981 | 0.29 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr5_+_140027355 | 0.28 |
ENST00000417647.2 ENST00000507593.1 ENST00000508301.1 |
IK |
IK cytokine, down-regulator of HLA II |
| chr4_-_99850243 | 0.27 |
ENST00000280892.6 ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr12_-_85306594 | 0.24 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr1_-_161207986 | 0.22 |
ENST00000506209.1 ENST00000367980.2 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr12_-_110888103 | 0.22 |
ENST00000426440.1 ENST00000228825.7 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr2_-_26205340 | 0.22 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr3_-_47950745 | 0.22 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr1_-_161208013 | 0.21 |
ENST00000515452.1 ENST00000367983.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_19988566 | 0.20 |
ENST00000273047.4 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr10_-_70092671 | 0.19 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr1_-_43855444 | 0.18 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr16_+_532503 | 0.17 |
ENST00000412256.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr6_-_56507586 | 0.16 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr4_+_170581213 | 0.15 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr5_-_88178964 | 0.14 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_+_113033164 | 0.14 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr4_-_164534657 | 0.14 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr10_-_105845674 | 0.13 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr2_-_228497888 | 0.13 |
ENST00000264387.4 ENST00000409066.1 |
C2orf83 |
chromosome 2 open reading frame 83 |
| chr2_-_169746878 | 0.13 |
ENST00000282074.2 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
| chr12_+_16109519 | 0.13 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr2_+_201980827 | 0.13 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr14_+_89060739 | 0.12 |
ENST00000318308.6 |
ZC3H14 |
zinc finger CCCH-type containing 14 |
| chr1_+_43855560 | 0.11 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr12_+_56324933 | 0.11 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr5_-_88179017 | 0.10 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr21_-_38445297 | 0.09 |
ENST00000430792.1 ENST00000399103.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr21_-_38445011 | 0.09 |
ENST00000464265.1 ENST00000399102.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr17_+_67498538 | 0.08 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr1_-_27701307 | 0.08 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr11_-_10590238 | 0.07 |
ENST00000256178.3 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr1_-_115238207 | 0.07 |
ENST00000520113.2 ENST00000369538.3 ENST00000353928.6 |
AMPD1 |
adenosine monophosphate deaminase 1 |
| chr19_+_13134772 | 0.07 |
ENST00000587760.1 ENST00000585575.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_-_101719159 | 0.07 |
ENST00000520868.1 ENST00000522658.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr15_+_80364901 | 0.07 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr7_-_74267836 | 0.07 |
ENST00000361071.5 ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2 |
GTF2I repeat domain containing 2 |
| chrX_-_65259914 | 0.07 |
ENST00000374737.4 ENST00000455586.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr11_-_107590383 | 0.06 |
ENST00000525934.1 ENST00000531293.1 |
SLN |
sarcolipin |
| chr5_-_1882858 | 0.06 |
ENST00000511126.1 ENST00000231357.2 |
IRX4 |
iroquois homeobox 4 |
| chr1_-_216978709 | 0.06 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr11_+_98891797 | 0.06 |
ENST00000527185.1 ENST00000528682.1 ENST00000524871.1 |
CNTN5 |
contactin 5 |
| chrX_-_65259900 | 0.05 |
ENST00000412866.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr14_+_65878565 | 0.05 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr7_+_74508372 | 0.04 |
ENST00000356115.5 ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B |
GTF2I repeat domain containing 2B |
| chr13_+_49551020 | 0.04 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr18_-_60986613 | 0.03 |
ENST00000444484.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr11_+_64004888 | 0.03 |
ENST00000541681.1 |
VEGFB |
vascular endothelial growth factor B |
| chr4_-_48136217 | 0.03 |
ENST00000264316.4 |
TXK |
TXK tyrosine kinase |
| chr7_+_129710350 | 0.02 |
ENST00000335420.5 ENST00000463413.1 |
KLHDC10 |
kelch domain containing 10 |
| chr2_-_183291741 | 0.02 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr10_+_51187938 | 0.02 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chr7_+_141811539 | 0.02 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr11_+_120207787 | 0.02 |
ENST00000397843.2 ENST00000356641.3 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_181452678 | 0.02 |
ENST00000367570.1 ENST00000526775.1 ENST00000357570.5 ENST00000358338.5 ENST00000367567.4 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr2_+_181845843 | 0.01 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr14_-_24711865 | 0.00 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 3.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 10.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.7 | 2.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.7 | 2.2 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 3.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 5.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 0.5 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.2 | 0.5 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.5 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 2.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.5 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.9 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.5 | GO:0031293 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 1.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 4.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.7 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 5.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 2.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.4 | 2.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 3.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 2.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 4.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |