Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
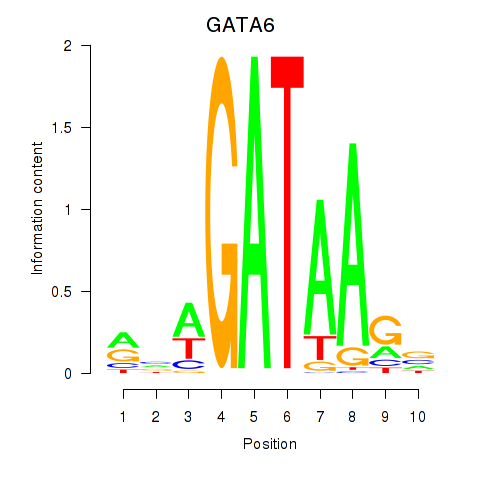
Results for GATA6
Z-value: 1.00
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.4 | GATA6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA6 | hg19_v2_chr18_+_19749386_19749404 | 0.20 | 4.5e-01 | Click! |
Activity profile of GATA6 motif
Sorted Z-values of GATA6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_62084241 | 4.01 |
ENST00000449662.2 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr16_+_202686 | 3.34 |
ENST00000252951.2 |
HBZ |
hemoglobin, zeta |
| chr12_-_54689532 | 3.20 |
ENST00000540264.2 ENST00000312156.4 |
NFE2 |
nuclear factor, erythroid 2 |
| chr19_-_13213662 | 2.62 |
ENST00000264824.4 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
| chr19_+_10397621 | 2.17 |
ENST00000380770.3 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr19_+_10397648 | 2.15 |
ENST00000340992.4 ENST00000393717.2 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr7_-_100239132 | 2.04 |
ENST00000223051.3 ENST00000431692.1 |
TFR2 |
transferrin receptor 2 |
| chr11_-_116708302 | 1.60 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr11_+_118958689 | 1.51 |
ENST00000535253.1 ENST00000392841.1 |
HMBS |
hydroxymethylbilane synthase |
| chr2_+_102928009 | 1.43 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr13_-_46716969 | 1.40 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr8_-_38008783 | 1.38 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr12_-_6233828 | 1.20 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr1_-_158656488 | 1.20 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr15_-_64126084 | 1.17 |
ENST00000560316.1 ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr8_+_21823726 | 1.16 |
ENST00000433566.4 |
XPO7 |
exportin 7 |
| chr11_-_66206260 | 1.11 |
ENST00000329819.4 ENST00000310999.7 ENST00000430466.2 |
MRPL11 |
mitochondrial ribosomal protein L11 |
| chr3_+_69812877 | 1.04 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_25598989 | 1.04 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chr17_+_74381343 | 1.03 |
ENST00000392496.3 |
SPHK1 |
sphingosine kinase 1 |
| chr2_+_3642545 | 0.97 |
ENST00000382062.2 ENST00000236693.7 ENST00000349077.4 |
COLEC11 |
collectin sub-family member 11 |
| chr1_+_161136180 | 0.94 |
ENST00000352210.5 ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX |
protoporphyrinogen oxidase |
| chr2_+_138722028 | 0.93 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr1_-_27240455 | 0.76 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr4_+_100495864 | 0.69 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr18_+_19749386 | 0.69 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr21_-_39870339 | 0.67 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr1_+_25598872 | 0.61 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr1_-_24469602 | 0.61 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr3_+_148583043 | 0.55 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr8_-_95449155 | 0.55 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr1_+_25599018 | 0.54 |
ENST00000417538.2 ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD |
Rh blood group, D antigen |
| chr2_-_44065889 | 0.53 |
ENST00000543989.1 ENST00000405322.1 |
ABCG5 |
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr17_+_4675175 | 0.53 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr4_-_103266355 | 0.53 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr2_-_44065946 | 0.51 |
ENST00000260645.1 |
ABCG5 |
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr1_+_12524965 | 0.50 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr20_-_30795511 | 0.46 |
ENST00000246229.4 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
| chr18_-_28742813 | 0.45 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr6_+_12290586 | 0.38 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr19_-_11494975 | 0.37 |
ENST00000222139.6 ENST00000592375.2 |
EPOR |
erythropoietin receptor |
| chr20_-_54967187 | 0.33 |
ENST00000422322.1 ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA |
aurora kinase A |
| chr5_+_159848807 | 0.32 |
ENST00000352433.5 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr19_-_55549624 | 0.30 |
ENST00000417454.1 ENST00000310373.3 ENST00000333884.2 |
GP6 |
glycoprotein VI (platelet) |
| chr14_+_75988851 | 0.29 |
ENST00000555504.1 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr6_-_30128657 | 0.28 |
ENST00000449742.2 ENST00000376704.3 |
TRIM10 |
tripartite motif containing 10 |
| chr12_+_53818855 | 0.26 |
ENST00000550839.1 |
AMHR2 |
anti-Mullerian hormone receptor, type II |
| chr12_+_111843749 | 0.25 |
ENST00000341259.2 |
SH2B3 |
SH2B adaptor protein 3 |
| chr7_+_155090271 | 0.24 |
ENST00000476756.1 |
INSIG1 |
insulin induced gene 1 |
| chr2_-_219850277 | 0.23 |
ENST00000295727.1 |
FEV |
FEV (ETS oncogene family) |
| chr4_-_103266219 | 0.21 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr17_-_4806369 | 0.21 |
ENST00000293780.4 |
CHRNE |
cholinergic receptor, nicotinic, epsilon (muscle) |
| chr2_+_169757750 | 0.20 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr10_+_118350468 | 0.19 |
ENST00000358834.4 ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr3_+_69812701 | 0.19 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_86099898 | 0.19 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr4_-_186697044 | 0.18 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr9_-_117692697 | 0.17 |
ENST00000223795.2 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
| chr2_+_220436917 | 0.16 |
ENST00000243786.2 |
INHA |
inhibin, alpha |
| chr14_+_53173910 | 0.16 |
ENST00000606149.1 ENST00000555339.1 ENST00000556813.1 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr1_+_171217677 | 0.16 |
ENST00000402921.2 |
FMO1 |
flavin containing monooxygenase 1 |
| chr15_-_43513187 | 0.15 |
ENST00000540029.1 ENST00000441366.2 |
EPB42 |
erythrocyte membrane protein band 4.2 |
| chr16_+_71560154 | 0.15 |
ENST00000539698.3 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr10_+_118350522 | 0.15 |
ENST00000530319.1 ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr14_+_53173890 | 0.13 |
ENST00000445930.2 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr11_-_47374246 | 0.13 |
ENST00000545968.1 ENST00000399249.2 ENST00000256993.4 |
MYBPC3 |
myosin binding protein C, cardiac |
| chr5_-_33984741 | 0.12 |
ENST00000382102.3 ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr1_+_50574585 | 0.10 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr18_-_45663666 | 0.09 |
ENST00000535628.2 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
| chr20_+_43029911 | 0.08 |
ENST00000443598.2 ENST00000316099.4 ENST00000415691.2 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr2_-_214014959 | 0.07 |
ENST00000442445.1 ENST00000457361.1 ENST00000342002.2 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr5_-_33984786 | 0.07 |
ENST00000296589.4 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr7_-_107880508 | 0.06 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr1_+_171217622 | 0.05 |
ENST00000433267.1 ENST00000367750.3 |
FMO1 |
flavin containing monooxygenase 1 |
| chr16_+_71560023 | 0.04 |
ENST00000572450.1 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr8_-_20040638 | 0.03 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr14_+_21945335 | 0.03 |
ENST00000262709.3 ENST00000457430.2 ENST00000448790.2 |
TOX4 |
TOX high mobility group box family member 4 |
| chr8_-_93107443 | 0.02 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr14_+_75988768 | 0.01 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr14_-_21945057 | 0.01 |
ENST00000397762.1 |
RAB2B |
RAB2B, member RAS oncogene family |
| chr2_+_87565634 | 0.01 |
ENST00000421835.2 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr2_-_90538397 | 0.00 |
ENST00000443397.3 |
RP11-685N3.1 |
Uncharacterized protein |
| chr9_+_125796806 | 0.00 |
ENST00000373642.1 |
GPR21 |
G protein-coupled receptor 21 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 3.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 4.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 0.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 3.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 2.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 9.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 2.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.4 | 2.6 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 2.0 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 3.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 2.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 0.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.4 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.7 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.4 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 3.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.3 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 1.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 1.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 2.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.3 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.6 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 3.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 8.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 4.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |