Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
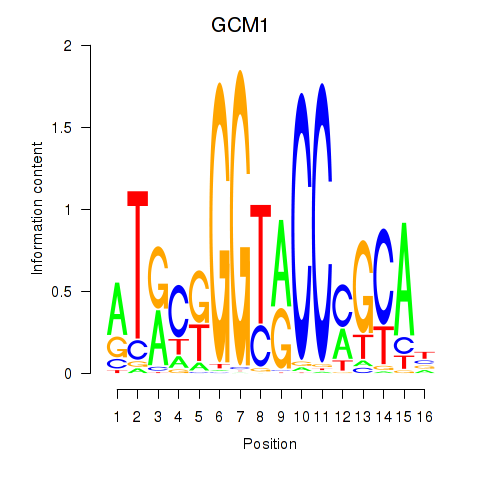
Results for GCM1
Z-value: 0.78
Transcription factors associated with GCM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM1
|
ENSG00000137270.10 | GCM1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM1 | hg19_v2_chr6_-_53013620_53013644 | -0.17 | 5.2e-01 | Click! |
Activity profile of GCM1 motif
Sorted Z-values of GCM1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23077065 | 2.63 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr22_+_22930626 | 2.47 |
ENST00000390302.2 |
IGLV2-33 |
immunoglobulin lambda variable 2-33 (non-functional) |
| chr22_+_23040274 | 2.12 |
ENST00000390306.2 |
IGLV2-23 |
immunoglobulin lambda variable 2-23 |
| chr22_+_23165153 | 1.96 |
ENST00000390317.2 |
IGLV2-8 |
immunoglobulin lambda variable 2-8 |
| chr18_+_21269556 | 1.61 |
ENST00000399516.3 |
LAMA3 |
laminin, alpha 3 |
| chr22_+_23101182 | 1.56 |
ENST00000390312.2 |
IGLV2-14 |
immunoglobulin lambda variable 2-14 |
| chr18_+_21529811 | 1.42 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21269404 | 1.29 |
ENST00000313654.9 |
LAMA3 |
laminin, alpha 3 |
| chr20_+_44441304 | 1.20 |
ENST00000352551.5 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr1_+_233765353 | 1.03 |
ENST00000366620.1 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr11_+_35198118 | 0.99 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr20_+_30639991 | 0.98 |
ENST00000534862.1 ENST00000538448.1 ENST00000375862.2 |
HCK |
hemopoietic cell kinase |
| chr1_+_158975744 | 0.94 |
ENST00000426592.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr10_+_131265443 | 0.82 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr5_-_142780280 | 0.75 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr10_-_105677427 | 0.74 |
ENST00000369764.1 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr22_+_22764088 | 0.70 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr9_-_35650900 | 0.68 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr15_-_60884706 | 0.64 |
ENST00000449337.2 |
RORA |
RAR-related orphan receptor A |
| chr17_-_3599327 | 0.63 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr14_-_23285069 | 0.63 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr8_+_144099896 | 0.61 |
ENST00000292494.6 ENST00000429120.2 |
LY6E |
lymphocyte antigen 6 complex, locus E |
| chr8_+_144099914 | 0.60 |
ENST00000521699.1 ENST00000520531.1 ENST00000520466.1 ENST00000521003.1 ENST00000522528.1 ENST00000522971.1 ENST00000519611.1 ENST00000521182.1 ENST00000519546.1 ENST00000523847.1 ENST00000522024.1 |
LY6E |
lymphocyte antigen 6 complex, locus E |
| chr7_-_93520259 | 0.59 |
ENST00000222543.5 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr22_+_22712087 | 0.58 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chr17_-_3599492 | 0.57 |
ENST00000435558.1 ENST00000345901.3 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr9_+_117373486 | 0.56 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr7_-_26904317 | 0.52 |
ENST00000345317.2 |
SKAP2 |
src kinase associated phosphoprotein 2 |
| chr5_-_41870621 | 0.51 |
ENST00000196371.5 |
OXCT1 |
3-oxoacid CoA transferase 1 |
| chr10_-_105677886 | 0.50 |
ENST00000224950.3 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr19_+_12902289 | 0.47 |
ENST00000302754.4 |
JUNB |
jun B proto-oncogene |
| chr12_+_11802753 | 0.44 |
ENST00000396373.4 |
ETV6 |
ets variant 6 |
| chr17_-_7493390 | 0.44 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr5_-_169725231 | 0.42 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr1_+_44399466 | 0.40 |
ENST00000498139.2 ENST00000491846.1 |
ARTN |
artemin |
| chr4_-_57547870 | 0.39 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr17_-_4269920 | 0.37 |
ENST00000572484.1 |
UBE2G1 |
ubiquitin-conjugating enzyme E2G 1 |
| chr14_+_65879437 | 0.37 |
ENST00000394585.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr4_-_48782259 | 0.37 |
ENST00000507711.1 ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL |
FRY-like |
| chr3_+_130612803 | 0.36 |
ENST00000510168.1 ENST00000508532.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_+_99971068 | 0.36 |
ENST00000198536.2 ENST00000453419.1 |
PILRA |
paired immunoglobin-like type 2 receptor alpha |
| chr11_-_108422926 | 0.35 |
ENST00000428840.1 ENST00000526312.1 |
EXPH5 |
exophilin 5 |
| chr3_+_130613001 | 0.34 |
ENST00000504948.1 ENST00000513801.1 ENST00000505072.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_12977513 | 0.34 |
ENST00000330881.5 |
PRAMEF7 |
PRAME family member 7 |
| chr11_+_19138670 | 0.32 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr4_-_57547454 | 0.31 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr11_-_8986474 | 0.31 |
ENST00000525069.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr6_+_110012462 | 0.31 |
ENST00000441478.2 ENST00000230124.3 |
FIG4 |
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
| chr5_-_137071756 | 0.31 |
ENST00000394937.3 ENST00000309755.4 |
KLHL3 |
kelch-like family member 3 |
| chr14_+_65878565 | 0.30 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr9_+_100263912 | 0.29 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chr17_-_4269768 | 0.28 |
ENST00000396981.2 |
UBE2G1 |
ubiquitin-conjugating enzyme E2G 1 |
| chr11_-_8986279 | 0.28 |
ENST00000534025.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr21_-_38445443 | 0.28 |
ENST00000360525.4 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr3_+_130613226 | 0.27 |
ENST00000509662.1 ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr11_-_8985927 | 0.27 |
ENST00000528117.1 ENST00000309134.5 |
TMEM9B |
TMEM9 domain family, member B |
| chr10_-_73976025 | 0.27 |
ENST00000342444.4 ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr10_-_73975657 | 0.27 |
ENST00000394919.1 ENST00000526751.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr21_-_45660723 | 0.26 |
ENST00000344330.4 ENST00000407780.3 ENST00000400379.3 |
ICOSLG |
inducible T-cell co-stimulator ligand |
| chr15_+_45422178 | 0.25 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr16_+_67063262 | 0.25 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr3_-_121468602 | 0.24 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr1_+_67773044 | 0.24 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr18_-_18691739 | 0.24 |
ENST00000399799.2 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
| chr11_+_20385327 | 0.24 |
ENST00000451739.2 ENST00000532505.1 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr11_+_20385666 | 0.22 |
ENST00000532081.1 ENST00000531058.1 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr15_+_45422131 | 0.22 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr3_-_121468513 | 0.22 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr6_-_97285336 | 0.22 |
ENST00000229955.3 ENST00000417980.1 |
GPR63 |
G protein-coupled receptor 63 |
| chr8_+_142138799 | 0.21 |
ENST00000518668.1 |
DENND3 |
DENN/MADD domain containing 3 |
| chr2_-_136875712 | 0.21 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr16_-_85722530 | 0.21 |
ENST00000253462.3 |
GINS2 |
GINS complex subunit 2 (Psf2 homolog) |
| chr5_-_79287060 | 0.20 |
ENST00000512560.1 ENST00000509852.1 ENST00000512528.1 |
MTX3 |
metaxin 3 |
| chrX_-_84634737 | 0.20 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chrX_-_153151586 | 0.18 |
ENST00000370060.1 ENST00000370055.1 ENST00000420165.1 |
L1CAM |
L1 cell adhesion molecule |
| chr16_+_56598961 | 0.18 |
ENST00000219162.3 |
MT4 |
metallothionein 4 |
| chr17_+_75372165 | 0.17 |
ENST00000427674.2 |
SEPT9 |
septin 9 |
| chr5_-_127674883 | 0.17 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr19_-_6379069 | 0.17 |
ENST00000597721.1 |
PSPN |
persephin |
| chr7_-_143966381 | 0.17 |
ENST00000487179.1 |
CTAGE8 |
CTAGE family, member 8 |
| chr5_+_176784837 | 0.17 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr2_-_89385283 | 0.16 |
ENST00000390252.2 |
IGKV3-15 |
immunoglobulin kappa variable 3-15 |
| chr12_-_57824739 | 0.16 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chr9_-_116102562 | 0.16 |
ENST00000374193.4 ENST00000465979.1 |
WDR31 |
WD repeat domain 31 |
| chr1_+_24069952 | 0.16 |
ENST00000609199.1 |
TCEB3 |
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr2_+_946543 | 0.16 |
ENST00000308624.5 ENST00000407292.1 |
SNTG2 |
syntrophin, gamma 2 |
| chr3_-_128369643 | 0.16 |
ENST00000296255.3 |
RPN1 |
ribophorin I |
| chr11_+_20385231 | 0.16 |
ENST00000530266.1 ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr1_-_2145620 | 0.15 |
ENST00000545087.1 |
AL590822.1 |
Uncharacterized protein |
| chr2_-_24583168 | 0.15 |
ENST00000361999.3 |
ITSN2 |
intersectin 2 |
| chr16_+_67062996 | 0.15 |
ENST00000561924.2 |
CBFB |
core-binding factor, beta subunit |
| chr9_-_116102530 | 0.15 |
ENST00000374195.3 ENST00000341761.4 |
WDR31 |
WD repeat domain 31 |
| chr7_+_23221438 | 0.14 |
ENST00000258742.5 |
NUPL2 |
nucleoporin like 2 |
| chr1_-_11907829 | 0.14 |
ENST00000376480.3 |
NPPA |
natriuretic peptide A |
| chr3_-_133380731 | 0.14 |
ENST00000260810.5 |
TOPBP1 |
topoisomerase (DNA) II binding protein 1 |
| chr22_+_46067678 | 0.14 |
ENST00000381061.4 ENST00000252934.5 |
ATXN10 |
ataxin 10 |
| chr4_-_119274121 | 0.14 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr4_+_76649797 | 0.13 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr7_+_143880559 | 0.13 |
ENST00000486333.1 |
CTAGE4 |
CTAGE family, member 4 |
| chr16_+_53468332 | 0.13 |
ENST00000262133.6 |
RBL2 |
retinoblastoma-like 2 (p130) |
| chr5_+_122847781 | 0.13 |
ENST00000395412.1 ENST00000395411.1 ENST00000345990.4 |
CSNK1G3 |
casein kinase 1, gamma 3 |
| chr11_-_62783303 | 0.13 |
ENST00000336232.2 ENST00000430500.2 |
SLC22A8 |
solute carrier family 22 (organic anion transporter), member 8 |
| chr15_-_83736091 | 0.13 |
ENST00000261721.4 |
BTBD1 |
BTB (POZ) domain containing 1 |
| chr10_-_134756030 | 0.13 |
ENST00000368586.5 ENST00000368582.2 |
TTC40 |
tetratricopeptide repeat domain 40 |
| chr19_-_12886327 | 0.12 |
ENST00000397668.3 ENST00000587178.1 ENST00000264827.5 |
HOOK2 |
hook microtubule-tethering protein 2 |
| chr1_-_8938736 | 0.12 |
ENST00000234590.4 |
ENO1 |
enolase 1, (alpha) |
| chr22_+_22936998 | 0.12 |
ENST00000390303.2 |
IGLV3-32 |
immunoglobulin lambda variable 3-32 (non-functional) |
| chr5_+_140767452 | 0.12 |
ENST00000519479.1 |
PCDHGB4 |
protocadherin gamma subfamily B, 4 |
| chr4_+_164415785 | 0.12 |
ENST00000513272.1 ENST00000513134.1 |
TMA16 |
translation machinery associated 16 homolog (S. cerevisiae) |
| chr11_+_17756279 | 0.12 |
ENST00000265969.6 |
KCNC1 |
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr6_+_42018251 | 0.12 |
ENST00000372978.3 ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8 |
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr19_-_38806560 | 0.11 |
ENST00000591755.1 ENST00000337679.8 ENST00000339413.6 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_-_62783276 | 0.11 |
ENST00000535878.1 ENST00000545207.1 |
SLC22A8 |
solute carrier family 22 (organic anion transporter), member 8 |
| chr19_+_5690207 | 0.11 |
ENST00000347512.3 |
RPL36 |
ribosomal protein L36 |
| chr5_+_140753444 | 0.10 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr6_-_56707943 | 0.10 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr1_+_161185032 | 0.10 |
ENST00000367992.3 ENST00000289902.1 |
FCER1G |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr16_+_2034183 | 0.10 |
ENST00000569451.1 ENST00000248114.6 ENST00000561710.1 |
GFER |
growth factor, augmenter of liver regeneration |
| chr7_-_44180673 | 0.09 |
ENST00000457314.1 ENST00000447951.1 ENST00000431007.1 |
MYL7 |
myosin, light chain 7, regulatory |
| chr19_+_50936142 | 0.09 |
ENST00000357701.5 |
MYBPC2 |
myosin binding protein C, fast type |
| chr19_+_641178 | 0.09 |
ENST00000166133.3 |
FGF22 |
fibroblast growth factor 22 |
| chr3_-_9994021 | 0.09 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr13_-_30169807 | 0.09 |
ENST00000380752.5 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr20_+_34043085 | 0.08 |
ENST00000397527.1 ENST00000342580.4 |
CEP250 |
centrosomal protein 250kDa |
| chr6_-_132032206 | 0.08 |
ENST00000314099.8 |
CTAGE9 |
CTAGE family, member 9 |
| chr3_+_38080691 | 0.08 |
ENST00000308059.6 ENST00000346219.3 ENST00000452631.2 |
DLEC1 |
deleted in lung and esophageal cancer 1 |
| chr17_+_15902694 | 0.08 |
ENST00000261647.5 ENST00000486880.2 |
TTC19 |
tetratricopeptide repeat domain 19 |
| chr15_-_23891175 | 0.08 |
ENST00000532292.1 |
MAGEL2 |
MAGE-like 2 |
| chr5_+_178322893 | 0.08 |
ENST00000361362.2 ENST00000520660.1 ENST00000520805.1 |
ZFP2 |
ZFP2 zinc finger protein |
| chr2_+_190649107 | 0.08 |
ENST00000441310.2 ENST00000409985.1 ENST00000446877.1 ENST00000418224.3 ENST00000409823.3 ENST00000374826.4 ENST00000424766.1 ENST00000447232.2 ENST00000432292.3 |
PMS1 |
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr11_+_124735282 | 0.08 |
ENST00000397801.1 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr11_-_100999775 | 0.08 |
ENST00000263463.5 |
PGR |
progesterone receptor |
| chr9_+_34958254 | 0.08 |
ENST00000242315.3 |
KIAA1045 |
KIAA1045 |
| chr16_+_333152 | 0.07 |
ENST00000219406.6 ENST00000404312.1 ENST00000456379.1 |
PDIA2 |
protein disulfide isomerase family A, member 2 |
| chr15_+_78632666 | 0.07 |
ENST00000299529.6 |
CRABP1 |
cellular retinoic acid binding protein 1 |
| chr17_-_15903002 | 0.07 |
ENST00000399277.1 |
ZSWIM7 |
zinc finger, SWIM-type containing 7 |
| chr14_+_24540731 | 0.07 |
ENST00000558859.1 ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6 |
copine VI (neuronal) |
| chr12_-_104443890 | 0.06 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr1_-_8483723 | 0.06 |
ENST00000476556.1 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
| chr3_+_192958914 | 0.06 |
ENST00000264735.2 ENST00000602513.1 |
HRASLS |
HRAS-like suppressor |
| chr12_-_4554780 | 0.06 |
ENST00000228837.2 |
FGF6 |
fibroblast growth factor 6 |
| chr11_-_8190534 | 0.06 |
ENST00000309737.6 ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
| chr1_+_25071848 | 0.06 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr7_-_4901625 | 0.06 |
ENST00000404991.1 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
| chr4_+_39408470 | 0.06 |
ENST00000257408.4 |
KLB |
klotho beta |
| chr15_-_78933567 | 0.06 |
ENST00000261751.3 ENST00000412074.2 |
CHRNB4 |
cholinergic receptor, nicotinic, beta 4 (neuronal) |
| chr1_-_40237020 | 0.05 |
ENST00000327582.5 |
OXCT2 |
3-oxoacid CoA transferase 2 |
| chr12_+_69864129 | 0.05 |
ENST00000547219.1 ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
| chr14_+_24540046 | 0.05 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr6_-_136847610 | 0.05 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr9_-_136004782 | 0.05 |
ENST00000393157.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr3_-_10749696 | 0.05 |
ENST00000397077.1 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr19_+_50321528 | 0.05 |
ENST00000312865.6 ENST00000595185.1 ENST00000538643.1 |
MED25 |
mediator complex subunit 25 |
| chr5_+_134094461 | 0.05 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr16_-_21170762 | 0.05 |
ENST00000261383.3 ENST00000415178.1 |
DNAH3 |
dynein, axonemal, heavy chain 3 |
| chr6_+_33589161 | 0.05 |
ENST00000605930.1 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
| chrX_-_109590174 | 0.05 |
ENST00000372054.1 |
GNG5P2 |
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr16_+_2285817 | 0.04 |
ENST00000564065.1 |
DNASE1L2 |
deoxyribonuclease I-like 2 |
| chr9_-_111619239 | 0.04 |
ENST00000374667.3 |
ACTL7B |
actin-like 7B |
| chr21_+_35747773 | 0.04 |
ENST00000399292.3 ENST00000399299.1 ENST00000399295.2 |
SMIM11 |
small integral membrane protein 11 |
| chr19_+_11546440 | 0.04 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr4_-_52904425 | 0.04 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr4_+_110749143 | 0.04 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr7_-_44179972 | 0.04 |
ENST00000446581.1 |
MYL7 |
myosin, light chain 7, regulatory |
| chr8_+_141521386 | 0.04 |
ENST00000220913.5 ENST00000519533.1 |
CHRAC1 |
chromatin accessibility complex 1 |
| chr16_-_90096309 | 0.04 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr1_+_1215816 | 0.04 |
ENST00000379116.5 |
SCNN1D |
sodium channel, non-voltage-gated 1, delta subunit |
| chr7_-_50132860 | 0.03 |
ENST00000046087.2 |
ZPBP |
zona pellucida binding protein |
| chr8_+_145133493 | 0.03 |
ENST00000316052.5 ENST00000525936.1 |
EXOSC4 |
exosome component 4 |
| chr19_-_49121054 | 0.03 |
ENST00000546623.1 ENST00000084795.5 |
RPL18 |
ribosomal protein L18 |
| chr18_+_10526008 | 0.03 |
ENST00000542979.1 ENST00000322897.6 |
NAPG |
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr4_-_40859132 | 0.03 |
ENST00000543538.1 ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr6_-_30043539 | 0.03 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr19_-_52227221 | 0.03 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr2_+_233320827 | 0.03 |
ENST00000295463.3 |
ALPI |
alkaline phosphatase, intestinal |
| chr7_-_50132801 | 0.03 |
ENST00000419417.1 |
ZPBP |
zona pellucida binding protein |
| chr1_+_45805342 | 0.02 |
ENST00000372090.5 |
TOE1 |
target of EGR1, member 1 (nuclear) |
| chr14_+_50779029 | 0.02 |
ENST00000245448.6 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr3_-_46506563 | 0.02 |
ENST00000231751.4 |
LTF |
lactotransferrin |
| chr16_+_67063142 | 0.02 |
ENST00000412916.2 |
CBFB |
core-binding factor, beta subunit |
| chr2_+_234959376 | 0.02 |
ENST00000425558.1 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
| chr10_+_73975742 | 0.02 |
ENST00000299381.4 |
ANAPC16 |
anaphase promoting complex subunit 16 |
| chr16_+_88872176 | 0.01 |
ENST00000569140.1 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
| chr9_+_102668915 | 0.01 |
ENST00000259400.6 ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17 |
syntaxin 17 |
| chr16_-_58768177 | 0.01 |
ENST00000434819.2 ENST00000245206.5 |
GOT2 |
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr1_+_1215968 | 0.01 |
ENST00000338555.2 |
SCNN1D |
sodium channel, non-voltage-gated 1, delta subunit |
| chr2_+_88469835 | 0.01 |
ENST00000358591.2 ENST00000377254.3 ENST00000402102.1 ENST00000419759.1 ENST00000449349.1 ENST00000343544.4 |
THNSL2 |
threonine synthase-like 2 (S. cerevisiae) |
| chr14_-_54908043 | 0.01 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr1_+_154244987 | 0.00 |
ENST00000328703.7 ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1 |
HCLS1 associated protein X-1 |
| chr9_-_99540328 | 0.00 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr1_-_148347506 | 0.00 |
ENST00000369189.3 |
NBPF20 |
neuroblastoma breakpoint family, member 20 |
| chr7_+_148936732 | 0.00 |
ENST00000335870.2 |
ZNF212 |
zinc finger protein 212 |
| chr20_+_36946029 | 0.00 |
ENST00000417318.1 |
BPI |
bactericidal/permeability-increasing protein |
| chr17_-_7218631 | 0.00 |
ENST00000577040.2 ENST00000389167.5 ENST00000391950.3 |
GPS2 |
G protein pathway suppressor 2 |
| chr14_+_50779071 | 0.00 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr22_+_22676808 | 0.00 |
ENST00000390290.2 |
IGLV1-51 |
immunoglobulin lambda variable 1-51 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.8 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.6 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 8.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 3.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 0.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 1.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 1.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.4 | GO:0070318 | myoblast development(GO:0048627) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 1.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.4 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 7.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 1.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 4.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0003335 | corneocyte development(GO:0003335) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |