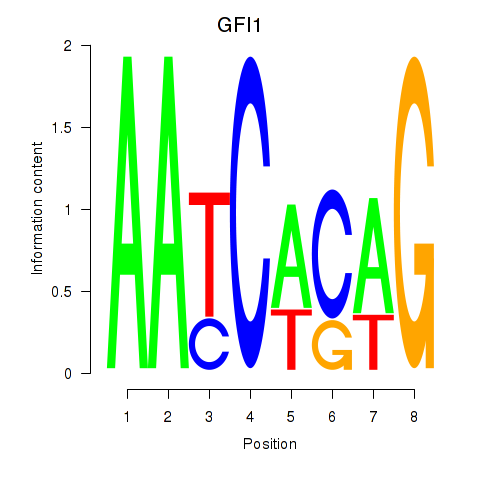
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for GFI1
Z-value: 0.91
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | GFI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92951607_92951661, hg19_v2_chr1_-_92949505_92949543, hg19_v2_chr1_-_92952433_92952489 | 0.62 | 1.0e-02 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47448102 | 2.59 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr4_-_155533787 | 2.45 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr2_+_11696464 | 2.34 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr17_-_26694979 | 2.30 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr17_-_26695013 | 2.23 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr14_-_22005062 | 1.91 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chr6_+_31982539 | 1.73 |
ENST00000435363.2 ENST00000425700.2 |
C4B |
complement component 4B (Chido blood group) |
| chr6_+_31949801 | 1.69 |
ENST00000428956.2 ENST00000498271.1 |
C4A |
complement component 4A (Rodgers blood group) |
| chr14_-_22005343 | 1.55 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr17_+_1665253 | 1.49 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chrX_+_49294472 | 1.48 |
ENST00000361446.5 |
GAGE12B |
G antigen 12B |
| chr17_-_7018128 | 1.41 |
ENST00000380952.2 ENST00000254850.7 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr17_-_7017968 | 1.40 |
ENST00000355035.5 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr17_+_1665345 | 1.37 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_-_145642267 | 1.35 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr11_+_22689648 | 1.24 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr8_-_145641864 | 1.20 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr17_+_4675175 | 1.10 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr17_-_79895097 | 1.06 |
ENST00000402252.2 ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr11_+_22688150 | 0.98 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr5_-_78809950 | 0.97 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr12_+_71833550 | 0.95 |
ENST00000266674.5 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr20_-_33999766 | 0.91 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr1_-_110306562 | 0.89 |
ENST00000369805.3 |
EPS8L3 |
EPS8-like 3 |
| chr7_-_87104963 | 0.89 |
ENST00000359206.3 ENST00000358400.3 ENST00000265723.4 |
ABCB4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr12_+_100867486 | 0.89 |
ENST00000548884.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr16_+_82090028 | 0.86 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr17_-_56595196 | 0.82 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr14_-_22005018 | 0.78 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr15_-_52587945 | 0.77 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr4_-_144940477 | 0.77 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr5_+_133451254 | 0.76 |
ENST00000517851.1 ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr10_-_82049424 | 0.73 |
ENST00000372213.3 |
MAT1A |
methionine adenosyltransferase I, alpha |
| chr4_-_145061788 | 0.73 |
ENST00000512064.1 ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA GYPB |
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr6_+_42984723 | 0.71 |
ENST00000332245.8 |
KLHDC3 |
kelch domain containing 3 |
| chr18_+_55711575 | 0.70 |
ENST00000356462.6 ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_54683419 | 0.69 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr6_+_42584847 | 0.68 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr10_-_13523073 | 0.68 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr13_-_46756351 | 0.68 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr10_+_94451574 | 0.65 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr10_-_11574274 | 0.64 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr17_-_79895154 | 0.63 |
ENST00000405481.4 ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr12_+_100867694 | 0.63 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr2_-_111291587 | 0.63 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr2_+_17721920 | 0.63 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr7_-_71912046 | 0.59 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr1_-_25291475 | 0.59 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr1_+_46713404 | 0.58 |
ENST00000371975.4 ENST00000469835.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr2_-_89157161 | 0.58 |
ENST00000390237.2 |
IGKC |
immunoglobulin kappa constant |
| chr1_+_46713357 | 0.57 |
ENST00000442598.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chrX_+_48681768 | 0.57 |
ENST00000430858.1 |
HDAC6 |
histone deacetylase 6 |
| chr19_-_13227534 | 0.56 |
ENST00000588229.1 ENST00000357720.4 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr6_-_31704282 | 0.55 |
ENST00000375784.3 ENST00000375779.2 |
CLIC1 |
chloride intracellular channel 1 |
| chr7_-_23510086 | 0.54 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr19_-_13227463 | 0.53 |
ENST00000437766.1 ENST00000221504.8 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr1_+_156338993 | 0.53 |
ENST00000368249.1 ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG |
Rh family, B glycoprotein (gene/pseudogene) |
| chr7_-_1980128 | 0.53 |
ENST00000437877.1 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr5_+_118690466 | 0.53 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chrX_-_48056199 | 0.52 |
ENST00000311798.1 ENST00000347757.1 |
SSX5 |
synovial sarcoma, X breakpoint 5 |
| chr16_-_70323422 | 0.52 |
ENST00000261772.8 |
AARS |
alanyl-tRNA synthetase |
| chr3_-_66024213 | 0.52 |
ENST00000483466.1 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_56494908 | 0.52 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr3_-_178976996 | 0.52 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr4_-_144826682 | 0.51 |
ENST00000358615.4 ENST00000437468.2 |
GYPE |
glycophorin E (MNS blood group) |
| chr16_+_12059050 | 0.51 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr17_-_56494882 | 0.51 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr16_+_12059091 | 0.51 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr2_-_96874553 | 0.50 |
ENST00000337288.5 ENST00000443962.1 |
STARD7 |
StAR-related lipid transfer (START) domain containing 7 |
| chr2_+_62900986 | 0.49 |
ENST00000405015.3 ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1 |
EH domain binding protein 1 |
| chr1_-_67142710 | 0.48 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr6_-_41040268 | 0.47 |
ENST00000373154.2 ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chrX_-_102319092 | 0.46 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr5_+_157158205 | 0.46 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr16_+_12058961 | 0.46 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_6772710 | 0.45 |
ENST00000304076.2 ENST00000602142.1 ENST00000596764.1 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
| chr6_+_45296048 | 0.45 |
ENST00000465038.2 ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr3_-_33686743 | 0.45 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr2_+_17721230 | 0.44 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chrX_-_52683950 | 0.44 |
ENST00000298181.5 |
SSX7 |
synovial sarcoma, X breakpoint 7 |
| chr17_-_56494713 | 0.42 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr14_-_95623607 | 0.42 |
ENST00000531162.1 ENST00000529720.1 ENST00000343455.3 |
DICER1 |
dicer 1, ribonuclease type III |
| chr2_-_27938593 | 0.42 |
ENST00000379677.2 |
AC074091.13 |
Uncharacterized protein |
| chr14_-_95624227 | 0.42 |
ENST00000526495.1 |
DICER1 |
dicer 1, ribonuclease type III |
| chr4_-_89744457 | 0.41 |
ENST00000395002.2 |
FAM13A |
family with sequence similarity 13, member A |
| chr1_-_173793246 | 0.41 |
ENST00000345664.6 ENST00000367710.3 |
CENPL |
centromere protein L |
| chrX_+_12885183 | 0.41 |
ENST00000380659.3 |
TLR7 |
toll-like receptor 7 |
| chr9_+_108456800 | 0.41 |
ENST00000434214.1 ENST00000374692.3 |
TMEM38B |
transmembrane protein 38B |
| chr1_-_173793458 | 0.41 |
ENST00000356198.2 |
CENPL |
centromere protein L |
| chr7_+_112063192 | 0.41 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr1_-_160681593 | 0.41 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr6_+_6588902 | 0.41 |
ENST00000230568.4 |
LY86 |
lymphocyte antigen 86 |
| chr8_+_40010989 | 0.41 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chr17_+_76311791 | 0.40 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr11_-_66675371 | 0.40 |
ENST00000393955.2 |
PC |
pyruvate carboxylase |
| chr10_-_65028938 | 0.40 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr11_+_117049854 | 0.40 |
ENST00000278951.7 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr2_+_47630108 | 0.39 |
ENST00000233146.2 ENST00000454849.1 ENST00000543555.1 |
MSH2 |
mutS homolog 2 |
| chr21_+_44313375 | 0.39 |
ENST00000354250.2 ENST00000340344.4 |
NDUFV3 |
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr17_-_18266660 | 0.39 |
ENST00000582653.1 ENST00000352886.6 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr6_-_25930904 | 0.38 |
ENST00000377850.3 |
SLC17A2 |
solute carrier family 17, member 2 |
| chr17_-_18266765 | 0.38 |
ENST00000354098.3 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr4_-_89744365 | 0.37 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr9_+_133710453 | 0.37 |
ENST00000318560.5 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr17_-_18266797 | 0.37 |
ENST00000316694.3 ENST00000539052.1 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr10_-_65028817 | 0.37 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr5_+_59783941 | 0.37 |
ENST00000506884.1 ENST00000504876.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr3_-_142166904 | 0.37 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr12_-_56756553 | 0.37 |
ENST00000398189.3 ENST00000541105.1 |
APOF |
apolipoprotein F |
| chr1_-_155947951 | 0.36 |
ENST00000313695.7 ENST00000497907.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_50353944 | 0.36 |
ENST00000594151.1 ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr8_+_21823726 | 0.36 |
ENST00000433566.4 |
XPO7 |
exportin 7 |
| chr3_+_51575596 | 0.36 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr1_+_32739733 | 0.36 |
ENST00000333070.4 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr7_-_148581251 | 0.36 |
ENST00000478654.1 ENST00000460911.1 ENST00000350995.2 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr20_+_36373032 | 0.36 |
ENST00000373473.1 |
CTNNBL1 |
catenin, beta like 1 |
| chrX_+_37639302 | 0.36 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr12_+_56732658 | 0.36 |
ENST00000228534.4 |
IL23A |
interleukin 23, alpha subunit p19 |
| chr11_-_72432950 | 0.36 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_-_41040195 | 0.36 |
ENST00000463088.1 ENST00000469104.1 ENST00000486443.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr7_+_55980331 | 0.35 |
ENST00000429591.2 |
ZNF713 |
zinc finger protein 713 |
| chr14_-_37642016 | 0.35 |
ENST00000331299.5 |
SLC25A21 |
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr7_+_130020932 | 0.35 |
ENST00000484324.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr7_+_100136811 | 0.35 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr3_+_63898275 | 0.35 |
ENST00000538065.1 |
ATXN7 |
ataxin 7 |
| chr12_-_56843161 | 0.34 |
ENST00000554616.1 ENST00000553532.1 ENST00000229201.4 |
TIMELESS |
timeless circadian clock |
| chr6_-_135271260 | 0.34 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr11_+_117049445 | 0.34 |
ENST00000324225.4 ENST00000532960.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr6_-_32920794 | 0.34 |
ENST00000395305.3 ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA XXbac-BPG181M17.5 |
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr8_-_27115903 | 0.33 |
ENST00000350889.4 ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4 |
stathmin-like 4 |
| chr5_+_59783540 | 0.33 |
ENST00000515734.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_152214098 | 0.33 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_153574480 | 0.33 |
ENST00000410080.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_-_89744314 | 0.32 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr6_+_167704838 | 0.32 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr4_+_41937131 | 0.32 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr12_+_117176113 | 0.32 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr8_-_27115931 | 0.31 |
ENST00000523048.1 |
STMN4 |
stathmin-like 4 |
| chr19_+_50354430 | 0.31 |
ENST00000599732.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr5_+_176784837 | 0.31 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr20_+_45338126 | 0.31 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr7_-_140482926 | 0.31 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr17_-_40761375 | 0.31 |
ENST00000543197.1 ENST00000309428.5 |
FAM134C |
family with sequence similarity 134, member C |
| chr20_-_34252759 | 0.31 |
ENST00000414711.1 ENST00000416778.1 ENST00000397442.1 ENST00000440240.1 ENST00000412056.1 ENST00000352393.4 ENST00000458038.1 ENST00000420363.1 ENST00000434795.1 ENST00000437100.1 ENST00000414664.1 ENST00000359646.1 ENST00000424458.1 ENST00000374104.3 ENST00000374114.3 |
CPNE1 RBM12 |
copine I RNA binding motif protein 12 |
| chr7_+_150758304 | 0.31 |
ENST00000482950.1 ENST00000463414.1 ENST00000310317.5 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr19_-_15529790 | 0.30 |
ENST00000596195.1 ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L |
A kinase (PRKA) anchor protein 8-like |
| chr1_+_168250194 | 0.30 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr3_+_52017454 | 0.30 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr19_+_50354462 | 0.30 |
ENST00000601675.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr5_-_147162078 | 0.30 |
ENST00000507386.1 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr2_-_68479614 | 0.29 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr2_-_238499725 | 0.29 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr4_+_106631966 | 0.29 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr2_-_158300556 | 0.29 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr10_-_74385811 | 0.28 |
ENST00000603011.1 ENST00000401998.3 ENST00000361114.5 ENST00000604238.1 |
MICU1 |
mitochondrial calcium uptake 1 |
| chr19_+_3880581 | 0.28 |
ENST00000450849.2 ENST00000301260.6 ENST00000398448.3 |
ATCAY |
ataxia, cerebellar, Cayman type |
| chr5_-_111091948 | 0.28 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr6_+_6588316 | 0.28 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr15_+_84115868 | 0.28 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr7_-_148581360 | 0.27 |
ENST00000320356.2 ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr18_-_19283649 | 0.27 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr11_-_72433346 | 0.27 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_+_111195973 | 0.27 |
ENST00000368885.3 ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1 |
adenosylmethionine decarboxylase 1 |
| chr16_+_31366536 | 0.26 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_+_171785012 | 0.26 |
ENST00000234160.4 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
| chr6_-_135271219 | 0.26 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr14_+_72052983 | 0.26 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr17_+_34171081 | 0.26 |
ENST00000585577.1 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr2_+_202122703 | 0.26 |
ENST00000447616.1 ENST00000358485.4 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
| chr10_+_31610064 | 0.25 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr6_-_32908792 | 0.25 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr7_+_123488124 | 0.25 |
ENST00000476325.1 |
HYAL4 |
hyaluronoglucosaminidase 4 |
| chr12_+_7941989 | 0.24 |
ENST00000229307.4 |
NANOG |
Nanog homeobox |
| chr19_-_16045220 | 0.24 |
ENST00000326742.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chrX_-_48216101 | 0.24 |
ENST00000298396.2 ENST00000376893.3 |
SSX3 |
synovial sarcoma, X breakpoint 3 |
| chr5_+_37379314 | 0.24 |
ENST00000265107.4 ENST00000504564.1 |
WDR70 |
WD repeat domain 70 |
| chr2_+_32853093 | 0.24 |
ENST00000448773.1 ENST00000317907.4 |
TTC27 |
tetratricopeptide repeat domain 27 |
| chr15_+_84116106 | 0.24 |
ENST00000535412.1 ENST00000324537.5 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr19_-_46088068 | 0.23 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr12_+_96252706 | 0.23 |
ENST00000266735.5 ENST00000553192.1 ENST00000552085.1 |
SNRPF |
small nuclear ribonucleoprotein polypeptide F |
| chr5_-_88179302 | 0.23 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr11_+_117049910 | 0.23 |
ENST00000431081.2 ENST00000524842.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr17_+_45728427 | 0.23 |
ENST00000540627.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr16_+_19183671 | 0.22 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr9_+_2159850 | 0.22 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_47813110 | 0.22 |
ENST00000355085.3 |
C5AR1 |
complement component 5a receptor 1 |
| chr17_-_3539558 | 0.21 |
ENST00000225519.3 |
SHPK |
sedoheptulokinase |
| chr1_+_12524965 | 0.21 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr16_-_67517716 | 0.21 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr8_+_75736761 | 0.21 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr19_+_50354393 | 0.20 |
ENST00000391842.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr6_+_28092338 | 0.20 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr19_-_51875523 | 0.20 |
ENST00000593572.1 ENST00000595157.1 |
NKG7 |
natural killer cell group 7 sequence |
| chr14_-_106069247 | 0.20 |
ENST00000479229.1 |
RP11-731F5.1 |
RP11-731F5.1 |
| chr8_+_145192672 | 0.20 |
ENST00000347708.4 |
FAM203A |
family with sequence similarity 203, member A |
| chr16_-_19729502 | 0.20 |
ENST00000219837.7 |
KNOP1 |
lysine-rich nucleolar protein 1 |
| chr14_+_35591858 | 0.20 |
ENST00000603544.1 |
KIAA0391 |
KIAA0391 |
| chr20_-_33732952 | 0.20 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_-_153573887 | 0.20 |
ENST00000493468.2 ENST00000545856.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr11_-_11374904 | 0.19 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.8 | 5.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.7 | 5.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.6 | 4.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.6 | 6.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.3 | 4.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 1.3 | 3.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.1 | 12.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.1 | 5.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.0 | 2.9 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.9 | 4.5 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.9 | 6.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 3.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 4.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 1.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.6 | 5.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.6 | 1.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.5 | 2.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 | 5.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.4 | 2.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 1.2 | GO:0032679 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.4 | 2.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 1.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 3.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 | 0.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 0.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 2.0 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.3 | 0.8 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.3 | 2.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 2.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 6.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 2.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 0.7 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.6 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 3.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.6 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 1.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 2.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.1 | 0.4 | GO:0045359 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 2.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.7 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 2.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) formation of anatomical boundary(GO:0048859) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 2.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 6.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 6.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 12.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 6.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 6.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 10.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 4.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.3 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 3.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 5.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 4.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.0 | 5.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.9 | 5.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 4.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 5.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 4.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 3.8 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 3.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 8.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 12.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.9 | GO:0046625 | sphingosine-1-phosphate receptor activity(GO:0038036) sphingolipid binding(GO:0046625) |
| 0.2 | 0.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 2.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 2.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 4.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.5 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 6.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 4.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 10.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.5 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 1.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 4.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 14.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 8.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0016831 | carbon-carbon lyase activity(GO:0016830) carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0031673 | H zone(GO:0031673) |
| 1.0 | 3.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 4.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 5.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.7 | 3.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 10.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 2.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 6.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 5.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 2.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 4.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 12.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 6.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 3.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 4.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) GID complex(GO:0034657) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
| 0.0 | 2.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 2.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 3.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 3.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 1.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 2.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.3 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.9 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 2.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.4 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.2 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.3 | GO:0016018 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 2.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 2.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 4.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.4 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 2.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 2.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 4.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.5 | 1.5 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 1.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.4 | 1.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.3 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 0.9 | GO:0097327 | response to antineoplastic agent(GO:0097327) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.3 | 1.7 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 0.3 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.3 | 0.8 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 1.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 3.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 2.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 2.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 0.7 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.2 | 0.6 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 0.6 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 | 0.8 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 0.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 | 0.8 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 2.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 1.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.7 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.1 | GO:1901657 | glycosyl compound metabolic process(GO:1901657) |
| 0.1 | 0.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 1.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.6 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 1.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.3 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.8 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.1 | 0.2 | GO:0035963 | cellular response to interleukin-13(GO:0035963) |
| 0.1 | 0.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.4 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.4 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0072719 | cellular response to hydroxyurea(GO:0072711) cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 2.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.1 | GO:0039003 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) kidney rudiment formation(GO:0072003) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:2001280 | maternal aggressive behavior(GO:0002125) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 1.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.4 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 2.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 13.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 6.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 6.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 5.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 7.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |