Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for GGCUCAG
Z-value: 0.81
miRNA associated with seed GGCUCAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-24-3p
|
MIMAT0000080 |
Activity profile of GGCUCAG motif
Sorted Z-values of GGCUCAG motif
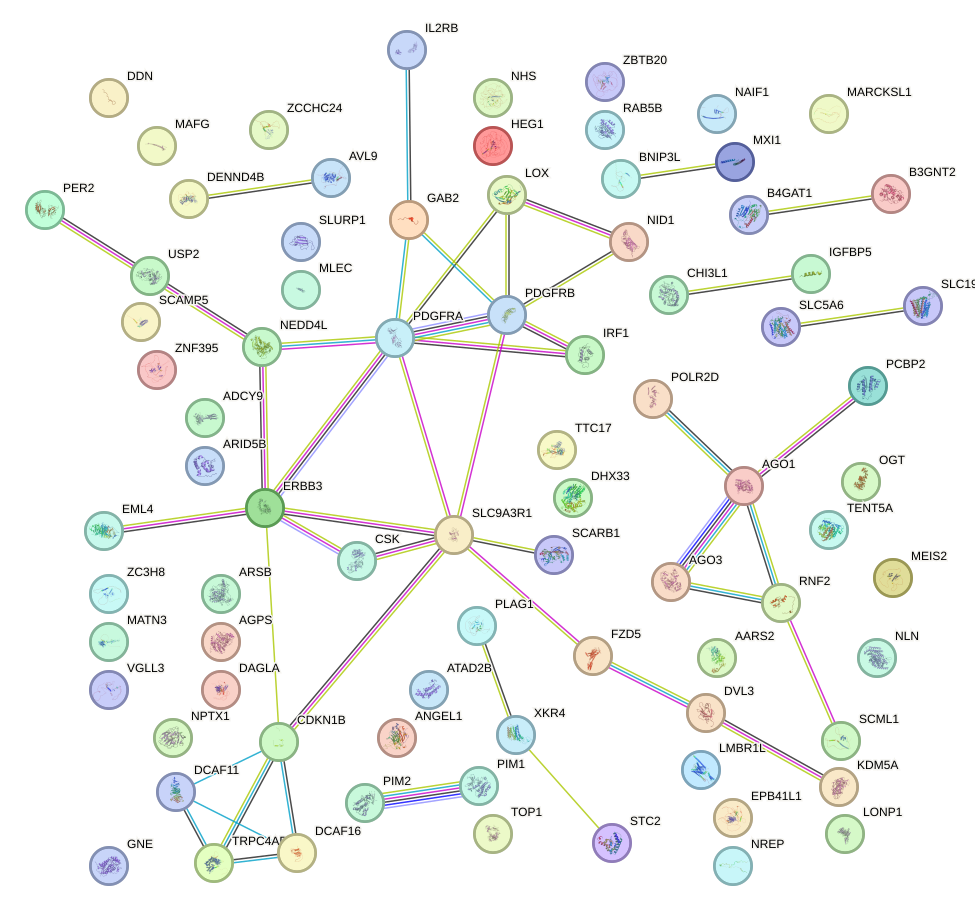
Network of associatons between targets according to the STRING database.
First level regulatory network of GGCUCAG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95360983 | 1.55 |
ENST00000371464.3 |
RBP4 |
retinol binding protein 4, plasma |
| chr10_+_63661053 | 1.32 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr5_-_172756506 | 1.28 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr1_-_236228403 | 1.13 |
ENST00000366595.3 |
NID1 |
nidogen 1 |
| chr5_-_111093406 | 1.10 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr12_+_12870055 | 1.09 |
ENST00000228872.4 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr8_-_28243934 | 0.87 |
ENST00000521185.1 ENST00000520290.1 ENST00000344423.5 |
ZNF395 |
zinc finger protein 395 |
| chr5_-_121413974 | 0.80 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr12_+_56473628 | 0.80 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_-_149535421 | 0.79 |
ENST00000261799.4 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
| chr1_-_169455169 | 0.77 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr20_-_52210368 | 0.73 |
ENST00000371471.2 |
ZNF217 |
zinc finger protein 217 |
| chr2_-_208634287 | 0.71 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chr1_-_32801825 | 0.67 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr2_-_69614373 | 0.61 |
ENST00000361060.5 ENST00000357308.4 |
GFPT1 |
glutamine--fructose-6-phosphate transaminase 1 |
| chr2_-_217560248 | 0.60 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr2_-_239197201 | 0.60 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr15_+_75287861 | 0.56 |
ENST00000425597.3 ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5 |
secretory carrier membrane protein 5 |
| chr11_-_78052923 | 0.56 |
ENST00000340149.2 |
GAB2 |
GRB2-associated binding protein 2 |
| chr11_-_66115032 | 0.56 |
ENST00000311181.4 |
B3GNT1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr9_-_130829588 | 0.53 |
ENST00000373078.4 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
| chr17_+_72744791 | 0.52 |
ENST00000583369.1 ENST00000262613.5 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
| chr6_+_37137939 | 0.52 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr16_+_69599861 | 0.50 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr1_+_214161272 | 0.49 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr3_-_87040233 | 0.48 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr2_-_24149977 | 0.45 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr3_-_124774802 | 0.45 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr10_-_81205373 | 0.45 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr5_-_78281603 | 0.45 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr10_+_111967345 | 0.45 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr5_-_131826457 | 0.45 |
ENST00000437654.1 ENST00000245414.4 |
IRF1 |
interferon regulatory factor 1 |
| chr14_+_24583836 | 0.43 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr22_+_21771656 | 0.42 |
ENST00000407464.2 |
HIC2 |
hypermethylated in cancer 2 |
| chr8_+_56014949 | 0.40 |
ENST00000327381.6 |
XKR4 |
XK, Kell blood group complex subunit-related family, member 4 |
| chr15_-_37390482 | 0.40 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr11_-_1593150 | 0.40 |
ENST00000397374.3 |
DUSP8 |
dual specificity phosphatase 8 |
| chrX_+_17755563 | 0.40 |
ENST00000380045.3 ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1 |
sex comb on midleg-like 1 (Drosophila) |
| chr5_+_65018017 | 0.40 |
ENST00000380985.5 ENST00000502464.1 |
NLN |
neurolysin (metallopeptidase M3 family) |
| chr20_+_39657454 | 0.38 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr17_+_34431212 | 0.37 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr4_+_55095264 | 0.37 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr20_+_34700333 | 0.37 |
ENST00000441639.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr15_+_75074385 | 0.36 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr12_-_125348448 | 0.36 |
ENST00000339570.5 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr15_-_34628951 | 0.34 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr12_-_498620 | 0.34 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr11_-_119252359 | 0.33 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr2_-_20212422 | 0.33 |
ENST00000421259.2 ENST00000407540.3 |
MATN3 |
matrilin 3 |
| chr8_+_26240414 | 0.33 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_+_36396677 | 0.32 |
ENST00000373191.4 ENST00000397828.2 |
AGO3 |
argonaute RISC catalytic component 3 |
| chr2_-_27435125 | 0.32 |
ENST00000414408.1 ENST00000310574.3 |
SLC5A6 |
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr12_-_49504655 | 0.31 |
ENST00000551782.1 ENST00000267102.8 |
LMBR1L |
limb development membrane protein 1-like |
| chr8_-_134584152 | 0.30 |
ENST00000521180.1 ENST00000517668.1 ENST00000319914.5 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_+_32535060 | 0.30 |
ENST00000318709.4 ENST00000409301.1 ENST00000404479.1 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
| chr1_-_115212696 | 0.30 |
ENST00000393276.3 ENST00000393277.1 |
DENND2C |
DENN/MADD domain containing 2C |
| chr2_+_201170703 | 0.29 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr6_-_44281043 | 0.29 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr2_+_42396472 | 0.29 |
ENST00000318522.5 ENST00000402711.2 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr3_+_183873098 | 0.29 |
ENST00000313143.3 |
DVL3 |
dishevelled segment polarity protein 3 |
| chr11_+_61447845 | 0.28 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr8_+_22102626 | 0.28 |
ENST00000519237.1 ENST00000397802.4 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr12_+_121124599 | 0.27 |
ENST00000228506.3 |
MLEC |
malectin |
| chr16_-_4166186 | 0.26 |
ENST00000294016.3 |
ADCY9 |
adenylate cyclase 9 |
| chr9_-_36276966 | 0.26 |
ENST00000543356.2 ENST00000396594.3 |
GNE |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr1_-_153919128 | 0.26 |
ENST00000361217.4 |
DENND4B |
DENN/MADD domain containing 4B |
| chr2_-_113012592 | 0.24 |
ENST00000272570.5 ENST00000409573.2 |
ZC3H8 |
zinc finger CCCH-type containing 8 |
| chr3_-_114790179 | 0.24 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr22_-_37545972 | 0.24 |
ENST00000216223.5 |
IL2RB |
interleukin 2 receptor, beta |
| chrX_+_70752917 | 0.24 |
ENST00000373719.3 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr2_+_102759199 | 0.24 |
ENST00000409288.1 ENST00000410023.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr12_+_56367697 | 0.23 |
ENST00000553116.1 ENST00000360299.5 ENST00000548068.1 ENST00000549915.1 ENST00000551459.1 ENST00000448789.2 |
RAB5B |
RAB5B, member RAS oncogene family |
| chr1_+_55464600 | 0.23 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr20_-_33999766 | 0.23 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr1_+_185014496 | 0.23 |
ENST00000367510.3 |
RNF2 |
ring finger protein 2 |
| chr17_-_78450398 | 0.22 |
ENST00000306773.4 |
NPTX1 |
neuronal pentraxin I |
| chr18_+_55711575 | 0.22 |
ENST00000356462.6 ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr20_-_33680588 | 0.22 |
ENST00000451813.2 ENST00000432634.2 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr12_+_53848505 | 0.22 |
ENST00000552819.1 ENST00000455667.3 |
PCBP2 |
poly(rC) binding protein 2 |
| chr6_-_82462425 | 0.22 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chrX_-_48776292 | 0.22 |
ENST00000376509.4 |
PIM2 |
pim-2 oncogene |
| chrX_-_131352152 | 0.21 |
ENST00000342983.2 |
RAP2C |
RAP2C, member of RAS oncogene family |
| chr12_-_49393092 | 0.21 |
ENST00000421952.2 |
DDN |
dendrin |
| chr17_-_5372271 | 0.20 |
ENST00000225296.3 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr11_+_43380459 | 0.20 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr10_-_103454876 | 0.20 |
ENST00000331272.7 |
FBXW4 |
F-box and WD repeat domain containing 4 |
| chr17_-_79885576 | 0.19 |
ENST00000574686.1 ENST00000357736.4 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr4_-_17812309 | 0.19 |
ENST00000382247.1 ENST00000536863.1 |
DCAF16 |
DDB1 and CUL4 associated factor 16 |
| chr1_-_203155868 | 0.19 |
ENST00000255409.3 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr2_+_178257372 | 0.18 |
ENST00000264167.4 ENST00000409888.1 |
AGPS |
alkylglycerone phosphate synthase |
| chr2_+_204192942 | 0.18 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr8_-_57123815 | 0.18 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr1_+_112162381 | 0.18 |
ENST00000433097.1 ENST00000369709.3 ENST00000436150.2 |
RAP1A |
RAP1A, member of RAS oncogene family |
| chr7_+_102004322 | 0.18 |
ENST00000496391.1 |
PRKRIP1 |
PRKR interacting protein 1 (IL11 inducible) |
| chr5_+_137774706 | 0.18 |
ENST00000378339.2 ENST00000254901.5 ENST00000506158.1 |
REEP2 |
receptor accessory protein 2 |
| chr14_-_77279153 | 0.17 |
ENST00000251089.2 |
ANGEL1 |
angel homolog 1 (Drosophila) |
| chr1_+_36348790 | 0.17 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr8_-_37756972 | 0.17 |
ENST00000330843.4 ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
| chr3_-_178790057 | 0.17 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr2_-_175351744 | 0.17 |
ENST00000295500.4 ENST00000392552.2 ENST00000392551.2 |
GPR155 |
G protein-coupled receptor 155 |
| chr17_+_58677539 | 0.16 |
ENST00000305921.3 |
PPM1D |
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr9_+_114659046 | 0.16 |
ENST00000374279.3 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
| chr15_+_41851211 | 0.16 |
ENST00000263798.3 |
TYRO3 |
TYRO3 protein tyrosine kinase |
| chr17_-_61777459 | 0.16 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr1_+_182992545 | 0.16 |
ENST00000258341.4 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
| chr4_+_1795012 | 0.16 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr15_-_35261996 | 0.16 |
ENST00000156471.5 |
AQR |
aquarius intron-binding spliceosomal factor |
| chr20_+_34894247 | 0.16 |
ENST00000373913.3 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr22_-_42466782 | 0.16 |
ENST00000396398.3 ENST00000403363.1 ENST00000402937.1 |
NAGA |
N-acetylgalactosaminidase, alpha- |
| chr15_+_65134088 | 0.16 |
ENST00000323544.4 ENST00000437723.1 |
PLEKHO2 AC069368.3 |
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr16_+_66914264 | 0.16 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr20_+_35201857 | 0.16 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr19_-_47735918 | 0.16 |
ENST00000449228.1 ENST00000300880.7 ENST00000341983.4 |
BBC3 |
BCL2 binding component 3 |
| chr2_-_131850951 | 0.15 |
ENST00000409185.1 ENST00000389915.3 |
FAM168B |
family with sequence similarity 168, member B |
| chr16_+_725650 | 0.15 |
ENST00000352681.3 ENST00000561556.1 |
RHBDL1 |
rhomboid, veinlet-like 1 (Drosophila) |
| chr17_-_41739283 | 0.15 |
ENST00000393661.2 ENST00000318579.4 |
MEOX1 |
mesenchyme homeobox 1 |
| chr16_+_20912382 | 0.15 |
ENST00000396052.2 |
LYRM1 |
LYR motif containing 1 |
| chr1_+_156124162 | 0.15 |
ENST00000368282.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_201979645 | 0.15 |
ENST00000367284.5 ENST00000367283.3 |
ELF3 |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr3_+_16926441 | 0.15 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr17_+_64298944 | 0.14 |
ENST00000413366.3 |
PRKCA |
protein kinase C, alpha |
| chr1_-_225615599 | 0.14 |
ENST00000421383.1 ENST00000272163.4 |
LBR |
lamin B receptor |
| chr9_+_37753795 | 0.14 |
ENST00000377753.2 ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B |
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chr17_+_19281034 | 0.14 |
ENST00000308406.5 ENST00000299612.7 |
MAPK7 |
mitogen-activated protein kinase 7 |
| chr17_-_9929581 | 0.14 |
ENST00000437099.2 ENST00000396115.2 |
GAS7 |
growth arrest-specific 7 |
| chr13_-_28545276 | 0.13 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chr1_+_178995021 | 0.13 |
ENST00000263733.4 |
FAM20B |
family with sequence similarity 20, member B |
| chr5_+_142149955 | 0.13 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr17_+_1958388 | 0.13 |
ENST00000399849.3 |
HIC1 |
hypermethylated in cancer 1 |
| chr11_+_65837907 | 0.13 |
ENST00000320580.4 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
| chr5_-_115910630 | 0.13 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_133451254 | 0.12 |
ENST00000517851.1 ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_-_200992827 | 0.12 |
ENST00000332129.2 ENST00000422435.2 |
KIF21B |
kinesin family member 21B |
| chr17_+_46018872 | 0.12 |
ENST00000583599.1 ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO |
pyridoxamine 5'-phosphate oxidase |
| chr3_+_45636219 | 0.12 |
ENST00000273317.4 |
LIMD1 |
LIM domains containing 1 |
| chr6_-_90529418 | 0.12 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr9_-_131534160 | 0.12 |
ENST00000291900.2 |
ZER1 |
zyg-11 related, cell cycle regulator |
| chr10_-_128077024 | 0.12 |
ENST00000368679.4 ENST00000368676.4 ENST00000448723.1 |
ADAM12 |
ADAM metallopeptidase domain 12 |
| chr17_-_72889697 | 0.12 |
ENST00000310226.6 |
FADS6 |
fatty acid desaturase 6 |
| chr3_-_52804872 | 0.11 |
ENST00000535191.1 ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4 |
NIMA-related kinase 4 |
| chr6_-_34360413 | 0.11 |
ENST00000607016.1 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr1_+_33116743 | 0.11 |
ENST00000414241.3 ENST00000373493.5 |
RBBP4 |
retinoblastoma binding protein 4 |
| chr16_-_67840442 | 0.11 |
ENST00000536251.1 ENST00000448631.2 ENST00000602677.1 ENST00000411657.2 ENST00000425512.2 ENST00000317506.3 |
RANBP10 |
RAN binding protein 10 |
| chr12_+_74931551 | 0.11 |
ENST00000519948.2 |
ATXN7L3B |
ataxin 7-like 3B |
| chr3_+_51575596 | 0.11 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr14_-_77843390 | 0.10 |
ENST00000216468.7 |
TMED8 |
transmembrane emp24 protein transport domain containing 8 |
| chr10_+_111767720 | 0.10 |
ENST00000356080.4 ENST00000277900.8 |
ADD3 |
adducin 3 (gamma) |
| chr22_-_51066521 | 0.10 |
ENST00000395621.3 ENST00000395619.3 ENST00000356098.5 ENST00000216124.5 ENST00000453344.2 ENST00000547307.1 ENST00000547805.1 |
ARSA |
arylsulfatase A |
| chrX_-_100548045 | 0.09 |
ENST00000372907.3 ENST00000372905.2 |
TAF7L |
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr17_+_12692774 | 0.09 |
ENST00000379672.5 ENST00000340825.3 |
ARHGAP44 |
Rho GTPase activating protein 44 |
| chr2_+_73441350 | 0.09 |
ENST00000389501.4 |
SMYD5 |
SMYD family member 5 |
| chr8_-_10588010 | 0.09 |
ENST00000304501.1 |
SOX7 |
SRY (sex determining region Y)-box 7 |
| chr13_+_98086445 | 0.09 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr3_+_49591881 | 0.09 |
ENST00000296452.4 |
BSN |
bassoon presynaptic cytomatrix protein |
| chr3_+_183353356 | 0.09 |
ENST00000242810.6 ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24 |
kelch-like family member 24 |
| chr1_+_215740709 | 0.08 |
ENST00000259154.4 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
| chr16_-_87525651 | 0.08 |
ENST00000268616.4 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
| chr20_+_32581452 | 0.08 |
ENST00000375114.3 ENST00000448364.1 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
| chr11_-_61684962 | 0.08 |
ENST00000394836.2 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr10_+_76586348 | 0.08 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr10_+_102222798 | 0.08 |
ENST00000343737.5 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
| chr16_-_71758602 | 0.08 |
ENST00000568954.1 |
PHLPP2 |
PH domain and leucine rich repeat protein phosphatase 2 |
| chr10_+_102295616 | 0.08 |
ENST00000299163.6 |
HIF1AN |
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr1_+_15943995 | 0.08 |
ENST00000480945.1 |
DDI2 |
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr9_+_102668915 | 0.08 |
ENST00000259400.6 ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17 |
syntaxin 17 |
| chr5_-_9546180 | 0.08 |
ENST00000382496.5 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr9_-_127269661 | 0.08 |
ENST00000373588.4 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr6_+_106546808 | 0.08 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr20_+_25176318 | 0.07 |
ENST00000354989.5 ENST00000360031.2 ENST00000376652.4 ENST00000439162.1 ENST00000433417.1 ENST00000417467.1 ENST00000433259.2 ENST00000427553.1 ENST00000435520.1 ENST00000418890.1 |
ENTPD6 |
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr22_+_31742875 | 0.07 |
ENST00000504184.2 |
AC005003.1 |
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr5_-_140053152 | 0.07 |
ENST00000542735.1 |
DND1 |
DND microRNA-mediated repression inhibitor 1 |
| chr2_-_26101374 | 0.07 |
ENST00000435504.4 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
| chr17_-_60142609 | 0.07 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr11_-_10590238 | 0.07 |
ENST00000256178.3 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr12_+_124196865 | 0.06 |
ENST00000330342.3 |
ATP6V0A2 |
ATPase, H+ transporting, lysosomal V0 subunit a2 |
| chr11_-_108093329 | 0.06 |
ENST00000278612.8 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
| chr13_-_70682590 | 0.06 |
ENST00000377844.4 |
KLHL1 |
kelch-like family member 1 |
| chr9_+_91933407 | 0.06 |
ENST00000375807.3 ENST00000339901.4 |
SECISBP2 |
SECIS binding protein 2 |
| chr6_+_35995488 | 0.06 |
ENST00000229795.3 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr5_+_138629417 | 0.06 |
ENST00000510056.1 ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3 |
matrin 3 |
| chr19_+_45754505 | 0.06 |
ENST00000262891.4 ENST00000300843.4 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
| chr17_+_33307503 | 0.06 |
ENST00000378526.4 ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3 |
ligase III, DNA, ATP-dependent |
| chr3_-_182698381 | 0.06 |
ENST00000292782.4 |
DCUN1D1 |
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr10_+_93683519 | 0.06 |
ENST00000265990.6 |
BTAF1 |
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr8_+_24772455 | 0.06 |
ENST00000433454.2 |
NEFM |
neurofilament, medium polypeptide |
| chr3_-_123603137 | 0.05 |
ENST00000360304.3 ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK |
myosin light chain kinase |
| chr3_-_120068143 | 0.05 |
ENST00000295628.3 |
LRRC58 |
leucine rich repeat containing 58 |
| chr8_-_13372395 | 0.05 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr3_+_187930719 | 0.05 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr1_-_160254913 | 0.05 |
ENST00000440949.3 ENST00000368072.5 ENST00000608310.1 ENST00000556710.1 |
PEX19 DCAF8 DCAF8 |
peroxisomal biogenesis factor 19 DDB1 and CUL4 associated factor 8 DDB1- and CUL4-associated factor 8 |
| chr5_+_139493665 | 0.05 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr19_-_2456922 | 0.05 |
ENST00000582871.1 ENST00000325327.3 |
LMNB2 |
lamin B2 |
| chr11_+_123396528 | 0.05 |
ENST00000322282.7 ENST00000529750.1 |
GRAMD1B |
GRAM domain containing 1B |
| chr3_-_197476560 | 0.05 |
ENST00000273582.5 |
KIAA0226 |
KIAA0226 |
| chr17_-_42345487 | 0.05 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr19_+_34745442 | 0.05 |
ENST00000299505.6 ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355 |
KIAA0355 |
| chr18_+_43914159 | 0.05 |
ENST00000588679.1 ENST00000269439.7 ENST00000543885.1 |
RNF165 |
ring finger protein 165 |
| chr1_+_27114418 | 0.05 |
ENST00000078527.4 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr21_+_45285050 | 0.04 |
ENST00000291572.8 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr5_+_145826867 | 0.04 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 1.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 0.8 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.2 | 0.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.2 | 0.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 1.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.5 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.6 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.4 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.3 | GO:0099542 | retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.1 | 0.9 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0061110 | histone H3-T6 phosphorylation(GO:0035408) dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.5 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.3 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.6 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 0.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |