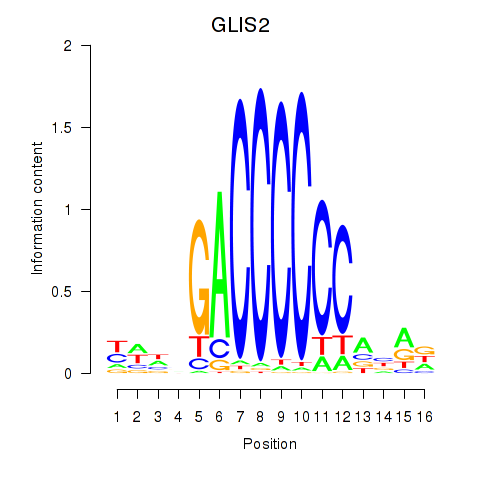
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for GLIS2
Z-value: 1.14
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS2 |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
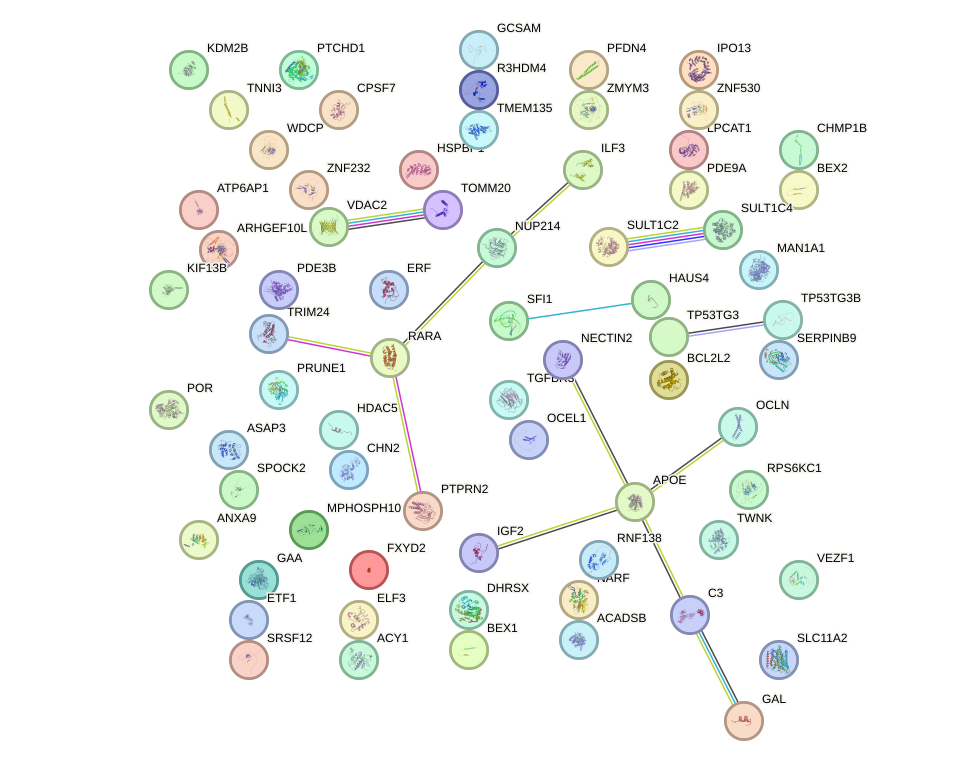
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_45418067 | 4.12 |
ENST00000589078.1 ENST00000586638.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_45417504 | 3.82 |
ENST00000588750.1 ENST00000588802.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_45417812 | 3.68 |
ENST00000592535.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_45417921 | 3.06 |
ENST00000252491.4 ENST00000592885.1 ENST00000589781.1 |
APOC1 |
apolipoprotein C-I |
| chr16_-_29910853 | 2.98 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr16_-_29910365 | 2.66 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr19_+_45409011 | 2.54 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr1_-_92371839 | 2.40 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr6_-_119670919 | 1.99 |
ENST00000368468.3 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
| chr7_+_75544466 | 1.92 |
ENST00000421059.1 ENST00000394893.1 ENST00000412521.1 ENST00000414186.1 |
POR |
P450 (cytochrome) oxidoreductase |
| chr7_+_75544397 | 1.89 |
ENST00000461988.1 ENST00000419840.1 |
POR |
P450 (cytochrome) oxidoreductase |
| chr19_-_6720686 | 1.72 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chrX_-_102319092 | 1.69 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr2_+_11752379 | 1.67 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr11_-_2160180 | 1.66 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr7_+_138145076 | 1.44 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr1_+_17866290 | 1.44 |
ENST00000361221.3 ENST00000452522.1 ENST00000434513.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_-_2903514 | 1.42 |
ENST00000380698.4 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
| chr1_-_19229248 | 1.32 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr11_+_22688150 | 1.17 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr17_+_79953310 | 1.11 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr14_-_21493884 | 0.98 |
ENST00000556974.1 ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2 |
NDRG family member 2 |
| chr17_-_42200996 | 0.98 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr22_+_21771656 | 0.91 |
ENST00000407464.2 |
HIC2 |
hypermethylated in cancer 2 |
| chr19_+_10527449 | 0.86 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr19_-_55669093 | 0.86 |
ENST00000344887.5 |
TNNI3 |
troponin I type 3 (cardiac) |
| chr12_-_51422017 | 0.84 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr11_+_14665263 | 0.83 |
ENST00000282096.4 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
| chr16_+_32264040 | 0.83 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chrX_+_51927919 | 0.81 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr19_+_45349432 | 0.81 |
ENST00000252485.4 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr17_-_56065484 | 0.79 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr16_-_32688053 | 0.79 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr3_+_157823609 | 0.78 |
ENST00000480820.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chrX_-_2418936 | 0.77 |
ENST00000461691.1 ENST00000381223.4 ENST00000381222.2 ENST00000412516.2 ENST00000334651.5 |
ZBED1 DHRSX |
zinc finger, BED-type containing 1 dehydrogenase/reductase (SDR family) X-linked |
| chr20_+_35974532 | 0.76 |
ENST00000373578.2 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr6_-_89827720 | 0.75 |
ENST00000452027.2 |
SRSF12 |
serine/arginine-rich splicing factor 12 |
| chrX_+_23352133 | 0.72 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr14_-_21493649 | 0.72 |
ENST00000553442.1 ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2 |
NDRG family member 2 |
| chr20_+_52824367 | 0.71 |
ENST00000371419.2 |
PFDN4 |
prefoldin subunit 4 |
| chr8_-_29120580 | 0.71 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr20_-_30310656 | 0.70 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr2_+_108905325 | 0.70 |
ENST00000438339.1 ENST00000409880.1 ENST00000437390.2 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr17_+_80416050 | 0.69 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr1_+_150954493 | 0.69 |
ENST00000368947.4 |
ANXA9 |
annexin A9 |
| chr17_+_78075361 | 0.68 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr5_+_68788594 | 0.64 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr19_+_782755 | 0.63 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr7_-_158380371 | 0.63 |
ENST00000389418.4 ENST00000389416.4 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr20_-_52210368 | 0.63 |
ENST00000371471.2 |
ZNF217 |
zinc finger protein 217 |
| chr22_-_21482352 | 0.58 |
ENST00000329949.3 |
POM121L7 |
POM121 transmembrane nucleoporin-like 7 |
| chr10_-_73848531 | 0.57 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr12_-_122018859 | 0.57 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr22_-_21213029 | 0.56 |
ENST00000572273.1 ENST00000255882.6 |
PI4KA |
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr9_+_134103496 | 0.55 |
ENST00000498010.1 ENST00000476004.1 ENST00000528406.1 |
NUP214 |
nucleoporin 214kDa |
| chr19_-_913160 | 0.54 |
ENST00000361574.5 ENST00000587975.1 |
R3HDM4 |
R3H domain containing 4 |
| chr10_+_124768482 | 0.53 |
ENST00000368869.4 ENST00000358776.4 |
ACADSB |
acyl-CoA dehydrogenase, short/branched chain |
| chr6_+_43044003 | 0.53 |
ENST00000230419.4 ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7 |
protein tyrosine kinase 7 |
| chr22_-_32026810 | 0.53 |
ENST00000266095.5 ENST00000397500.1 |
PISD |
phosphatidylserine decarboxylase |
| chr17_-_5015129 | 0.52 |
ENST00000575898.1 ENST00000416429.2 |
ZNF232 |
zinc finger protein 232 |
| chr1_+_201979645 | 0.52 |
ENST00000367284.5 ENST00000367283.3 |
ELF3 |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr20_-_30310693 | 0.50 |
ENST00000307677.4 ENST00000420653.1 |
BCL2L1 |
BCL2-like 1 |
| chr11_+_68451943 | 0.50 |
ENST00000265643.3 |
GAL |
galanin/GMAP prepropeptide |
| chr7_-_158380465 | 0.50 |
ENST00000389413.3 ENST00000409483.1 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr2_+_102508955 | 0.49 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chr19_+_58111241 | 0.48 |
ENST00000597700.1 ENST00000332854.6 ENST00000597864.1 |
ZNF530 |
zinc finger protein 530 |
| chr1_+_44412577 | 0.48 |
ENST00000372343.3 |
IPO13 |
importin 13 |
| chrX_-_70474910 | 0.47 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr20_-_23969416 | 0.47 |
ENST00000335694.4 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
| chr5_-_1524015 | 0.46 |
ENST00000283415.3 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
| chr11_+_86748863 | 0.46 |
ENST00000340353.7 |
TMEM135 |
transmembrane protein 135 |
| chr2_-_24270217 | 0.45 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chr10_+_114709999 | 0.45 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_-_55668093 | 0.44 |
ENST00000588882.1 ENST00000586858.1 |
TNNI3 |
troponin I type 3 (cardiac) |
| chr2_+_108905095 | 0.44 |
ENST00000251481.6 ENST00000326853.5 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_+_52017454 | 0.44 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr22_+_21400229 | 0.44 |
ENST00000342608.4 ENST00000543388.1 ENST00000442047.1 |
AC002472.13 |
Leucine-rich repeat-containing protein LOC400891 |
| chr17_+_38498594 | 0.43 |
ENST00000394081.3 |
RARA |
retinoic acid receptor, alpha |
| chr6_+_44214824 | 0.43 |
ENST00000371646.5 ENST00000353801.3 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr10_+_102747783 | 0.42 |
ENST00000311916.2 ENST00000370228.1 |
C10orf2 |
chromosome 10 open reading frame 2 |
| chr14_+_23776167 | 0.42 |
ENST00000554635.1 ENST00000557008.1 |
BCL2L2 BCL2L2-PABPN1 |
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr11_-_61197187 | 0.42 |
ENST00000449811.1 ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
| chr19_-_55791058 | 0.42 |
ENST00000587959.1 ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_+_65613217 | 0.41 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr16_+_33204980 | 0.40 |
ENST00000561509.1 |
TP53TG3C |
TP53 target 3C |
| chr1_+_150980889 | 0.40 |
ENST00000450884.1 ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE |
prune exopolyphosphatase |
| chr16_-_29415350 | 0.40 |
ENST00000524087.1 |
NPIPB11 |
nuclear pore complex interacting protein family, member B11 |
| chr3_-_48470838 | 0.39 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr1_+_213224572 | 0.39 |
ENST00000543470.1 ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr11_-_117695449 | 0.39 |
ENST00000292079.2 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr21_+_44073916 | 0.38 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr1_-_235292250 | 0.38 |
ENST00000366607.4 |
TOMM20 |
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr19_+_17337473 | 0.37 |
ENST00000598068.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr19_-_42759300 | 0.37 |
ENST00000222329.4 |
ERF |
Ets2 repressor factor |
| chr14_-_23426270 | 0.37 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr2_+_170655789 | 0.36 |
ENST00000409333.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr2_+_71357744 | 0.36 |
ENST00000498451.2 |
MPHOSPH10 |
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr7_+_29237354 | 0.36 |
ENST00000546235.1 |
CHN2 |
chimerin 2 |
| chr19_-_55791563 | 0.36 |
ENST00000588971.1 ENST00000255631.5 ENST00000587551.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_-_55791540 | 0.36 |
ENST00000433386.2 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_-_23810664 | 0.35 |
ENST00000336689.3 ENST00000437606.2 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chrX_+_153656978 | 0.35 |
ENST00000369762.2 ENST00000422890.1 |
ATP6AP1 |
ATPase, H+ transporting, lysosomal accessory protein 1 |
| chr19_+_17337406 | 0.35 |
ENST00000597836.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr19_-_3772209 | 0.34 |
ENST00000555978.1 ENST00000555633.1 |
RAX2 |
retina and anterior neural fold homeobox 2 |
| chr19_+_10765699 | 0.34 |
ENST00000590009.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr14_-_23426322 | 0.34 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chrX_+_48660287 | 0.33 |
ENST00000444343.2 ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6 |
histone deacetylase 6 |
| chr11_-_64510409 | 0.33 |
ENST00000394429.1 ENST00000394428.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_55791431 | 0.33 |
ENST00000593263.1 ENST00000376343.3 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr14_-_23426337 | 0.33 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_24615523 | 0.32 |
ENST00000559056.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_-_40169161 | 0.32 |
ENST00000589586.2 ENST00000426588.3 ENST00000589576.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr1_-_36947120 | 0.32 |
ENST00000361632.4 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr15_+_63796779 | 0.31 |
ENST00000561442.1 ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3 |
ubiquitin specific peptidase 3 |
| chrX_+_19373700 | 0.31 |
ENST00000379804.1 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr19_-_41859814 | 0.31 |
ENST00000221930.5 |
TGFB1 |
transforming growth factor, beta 1 |
| chr19_+_54372639 | 0.31 |
ENST00000391769.2 |
MYADM |
myeloid-associated differentiation marker |
| chr15_+_74610894 | 0.31 |
ENST00000558821.1 ENST00000268082.4 |
CCDC33 |
coiled-coil domain containing 33 |
| chr19_+_16999654 | 0.30 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr18_+_12948000 | 0.30 |
ENST00000585730.1 ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L |
SEH1-like (S. cerevisiae) |
| chr3_+_52719936 | 0.29 |
ENST00000418458.1 ENST00000394799.2 |
GNL3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_+_4535025 | 0.29 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_-_2702681 | 0.29 |
ENST00000382159.3 |
GNG7 |
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_109605663 | 0.29 |
ENST00000409271.1 ENST00000258443.2 ENST00000376651.1 |
EDAR |
ectodysplasin A receptor |
| chr16_+_69796209 | 0.29 |
ENST00000359154.2 ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr20_-_30310797 | 0.29 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr12_-_133263893 | 0.29 |
ENST00000535270.1 ENST00000320574.5 |
POLE |
polymerase (DNA directed), epsilon, catalytic subunit |
| chr19_+_13875316 | 0.29 |
ENST00000319545.8 ENST00000593245.1 ENST00000040663.6 |
MRI1 |
methylthioribose-1-phosphate isomerase 1 |
| chrX_+_118108571 | 0.28 |
ENST00000304778.7 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr16_-_29934558 | 0.28 |
ENST00000568995.1 ENST00000566413.1 |
KCTD13 |
potassium channel tetramerization domain containing 13 |
| chr20_-_35492048 | 0.28 |
ENST00000237536.4 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
| chr17_+_48503519 | 0.28 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr2_-_88285309 | 0.28 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr2_-_27886676 | 0.28 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr22_+_17082732 | 0.28 |
ENST00000558085.2 ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1 |
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr20_+_48807351 | 0.27 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr12_-_49110613 | 0.27 |
ENST00000261900.3 |
CCNT1 |
cyclin T1 |
| chr17_+_48503603 | 0.27 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr6_+_69345166 | 0.27 |
ENST00000370598.1 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr16_+_28505955 | 0.26 |
ENST00000564831.1 ENST00000328423.5 ENST00000431282.1 |
APOBR |
apolipoprotein B receptor |
| chr5_-_176836577 | 0.25 |
ENST00000253496.3 |
F12 |
coagulation factor XII (Hageman factor) |
| chr19_-_821931 | 0.25 |
ENST00000359894.2 ENST00000520876.3 ENST00000519502.1 |
LPPR3 |
hsa-mir-3187 |
| chr21_-_45671014 | 0.25 |
ENST00000436357.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr17_-_42345487 | 0.25 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr11_-_61197480 | 0.25 |
ENST00000439958.3 ENST00000394888.4 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
| chr17_+_7461580 | 0.24 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_-_18441131 | 0.24 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr19_-_41256207 | 0.24 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr8_+_67624653 | 0.24 |
ENST00000521198.2 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr4_+_40058411 | 0.24 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr22_-_50700140 | 0.24 |
ENST00000215659.8 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr3_+_180630444 | 0.23 |
ENST00000491062.1 ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
| chr16_+_30087288 | 0.23 |
ENST00000279387.7 ENST00000562664.1 ENST00000562222.1 |
PPP4C |
protein phosphatase 4, catalytic subunit |
| chr12_+_122242597 | 0.23 |
ENST00000267197.5 |
SETD1B |
SET domain containing 1B |
| chr1_+_2487800 | 0.23 |
ENST00000355716.4 |
TNFRSF14 |
tumor necrosis factor receptor superfamily, member 14 |
| chr12_-_95467356 | 0.23 |
ENST00000393101.3 ENST00000333003.5 |
NR2C1 |
nuclear receptor subfamily 2, group C, member 1 |
| chr14_-_24615805 | 0.23 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr16_+_69458428 | 0.22 |
ENST00000512062.1 ENST00000307892.8 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
| chr6_-_30815936 | 0.21 |
ENST00000442852.1 |
XXbac-BPG27H4.8 |
XXbac-BPG27H4.8 |
| chrX_+_118108601 | 0.21 |
ENST00000371628.3 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr3_+_4535155 | 0.21 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr15_-_68521996 | 0.21 |
ENST00000418702.2 ENST00000565471.1 ENST00000564752.1 ENST00000566347.1 ENST00000249806.5 ENST00000562767.1 |
CLN6 RP11-315D16.2 |
ceroid-lipofuscinosis, neuronal 6, late infantile, variant Uncharacterized protein |
| chr22_+_21213259 | 0.21 |
ENST00000215730.7 |
SNAP29 |
synaptosomal-associated protein, 29kDa |
| chr20_-_34638841 | 0.20 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chr4_-_48082192 | 0.20 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr12_-_6716569 | 0.20 |
ENST00000544040.1 ENST00000545942.1 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
| chr12_+_58166370 | 0.20 |
ENST00000300209.8 |
METTL21B |
methyltransferase like 21B |
| chr2_+_169312350 | 0.20 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr6_-_114292449 | 0.20 |
ENST00000519065.1 |
HDAC2 |
histone deacetylase 2 |
| chr19_-_16008880 | 0.20 |
ENST00000011989.7 ENST00000221700.6 |
CYP4F2 |
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr7_-_99679324 | 0.20 |
ENST00000292393.5 ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3 |
zinc finger protein 3 |
| chr19_+_1905365 | 0.20 |
ENST00000329478.2 ENST00000602400.1 ENST00000409472.1 |
ADAT3 SCAMP4 |
adenosine deaminase, tRNA-specific 3 secretory carrier membrane protein 4 |
| chr2_+_171785824 | 0.19 |
ENST00000452526.2 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
| chr17_-_5138099 | 0.19 |
ENST00000571800.1 ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
| chr17_-_4464081 | 0.19 |
ENST00000574154.1 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr14_+_58894141 | 0.19 |
ENST00000423743.3 |
KIAA0586 |
KIAA0586 |
| chr7_-_30739661 | 0.19 |
ENST00000445981.1 ENST00000348438.4 |
CRHR2 |
corticotropin releasing hormone receptor 2 |
| chr17_+_80014359 | 0.19 |
ENST00000578168.1 |
GPS1 |
G protein pathway suppressor 1 |
| chr15_+_52311398 | 0.19 |
ENST00000261845.5 |
MAPK6 |
mitogen-activated protein kinase 6 |
| chr7_+_130794846 | 0.18 |
ENST00000421797.2 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
| chr17_-_41132010 | 0.18 |
ENST00000409103.1 ENST00000360221.4 |
PTGES3L-AARSD1 |
PTGES3L-AARSD1 readthrough |
| chr19_-_19249255 | 0.18 |
ENST00000587583.2 ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A |
transmembrane protein 161A |
| chr11_-_111637083 | 0.18 |
ENST00000427203.2 ENST00000341980.6 ENST00000311129.5 ENST00000393055.2 ENST00000426998.2 ENST00000527614.1 |
PPP2R1B |
protein phosphatase 2, regulatory subunit A, beta |
| chr6_-_28367510 | 0.17 |
ENST00000361028.1 |
ZSCAN12 |
zinc finger and SCAN domain containing 12 |
| chr19_+_45582453 | 0.17 |
ENST00000591607.1 ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7 MARK4 |
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr11_-_46142615 | 0.17 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr19_+_49622646 | 0.17 |
ENST00000334186.4 |
PPFIA3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr2_+_131113609 | 0.17 |
ENST00000347849.3 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr15_+_91478493 | 0.17 |
ENST00000418476.2 |
UNC45A |
unc-45 homolog A (C. elegans) |
| chr3_-_125802765 | 0.17 |
ENST00000514891.1 ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr2_+_131113580 | 0.17 |
ENST00000175756.5 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr1_-_24126892 | 0.16 |
ENST00000374497.3 ENST00000425913.1 |
GALE |
UDP-galactose-4-epimerase |
| chr8_-_103136481 | 0.16 |
ENST00000524209.1 ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD |
neurocalcin delta |
| chr6_-_153304148 | 0.16 |
ENST00000229758.3 |
FBXO5 |
F-box protein 5 |
| chr9_+_87284675 | 0.16 |
ENST00000376208.1 ENST00000304053.6 ENST00000277120.3 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_-_627143 | 0.16 |
ENST00000176195.3 |
SCT |
secretin |
| chr1_+_91966384 | 0.16 |
ENST00000430031.2 ENST00000234626.6 |
CDC7 |
cell division cycle 7 |
| chr8_+_38243821 | 0.15 |
ENST00000519476.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_+_45458503 | 0.15 |
ENST00000337392.5 ENST00000591304.1 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
| chr9_+_117373486 | 0.15 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.3 | 3.8 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 0.8 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.3 | 2.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 1.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.8 | GO:0004803 | transposase activity(GO:0004803) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.5 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.8 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 1.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 1.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0015450 | protein channel activity(GO:0015266) P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.0 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) endopeptidase regulator activity(GO:0061135) |
| 0.0 | 0.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.7 | 14.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.6 | 2.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 1.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.6 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.2 | 0.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 1.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 3.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.7 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 1.3 | 3.8 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.8 | 2.5 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.8 | 2.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.6 | 1.7 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 1.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 2.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 1.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.2 | 1.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 1.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.8 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.2 | 0.6 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.2 | 0.6 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 1.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.7 | GO:0002086 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 0.2 | 0.8 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 0.5 | GO:0051795 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of catagen(GO:0051795) |
| 0.1 | 0.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.8 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.3 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.4 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.3 | GO:0060364 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) frontal suture morphogenesis(GO:0060364) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.2 | GO:0070201 | regulation of establishment of protein localization(GO:0070201) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 0.8 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.3 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.6 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 1.0 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 1.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1904303 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:1903949 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.0 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.0 | 0.3 | GO:1990440 | regulation of interleukin-6 biosynthetic process(GO:0045408) positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.6 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.6 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |