Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
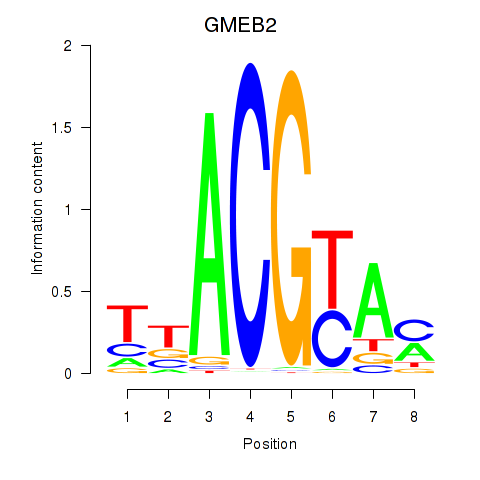
Results for GMEB2
Z-value: 1.54
Transcription factors associated with GMEB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB2
|
ENSG00000101216.6 | GMEB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | hg19_v2_chr20_-_62258394_62258464 | -0.23 | 4.0e-01 | Click! |
Activity profile of GMEB2 motif
Sorted Z-values of GMEB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_149106653 | 1.95 |
ENST00000462691.1 ENST00000370404.1 ENST00000483447.1 ENST00000370409.3 |
CXorf40B |
chromosome X open reading frame 40B |
| chr11_-_33913708 | 1.92 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chrX_+_48755183 | 1.87 |
ENST00000376563.1 ENST00000376566.4 |
PQBP1 |
polyglutamine binding protein 1 |
| chrX_+_48755202 | 1.86 |
ENST00000447146.2 ENST00000376548.5 ENST00000247140.4 |
PQBP1 |
polyglutamine binding protein 1 |
| chr7_+_120590803 | 1.76 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chrX_+_37545012 | 1.75 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr5_+_162887556 | 1.73 |
ENST00000393915.4 ENST00000432118.2 ENST00000358715.3 |
HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
| chr10_+_60145155 | 1.64 |
ENST00000373895.3 |
TFAM |
transcription factor A, mitochondrial |
| chr16_-_20911641 | 1.48 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr6_-_31763408 | 1.45 |
ENST00000444930.2 |
VARS |
valyl-tRNA synthetase |
| chr19_-_663277 | 1.42 |
ENST00000292363.5 |
RNF126 |
ring finger protein 126 |
| chr7_+_12726474 | 1.39 |
ENST00000396662.1 ENST00000356797.3 ENST00000396664.2 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr7_-_45151272 | 1.35 |
ENST00000461363.1 ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4 |
transforming growth factor beta regulator 4 |
| chr14_-_65346555 | 1.29 |
ENST00000542895.1 ENST00000556626.1 |
SPTB |
spectrin, beta, erythrocytic |
| chr15_+_66797455 | 1.24 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr18_+_2571510 | 1.22 |
ENST00000261597.4 ENST00000575515.1 |
NDC80 |
NDC80 kinetochore complex component |
| chr19_-_18548962 | 1.19 |
ENST00000317018.6 ENST00000581800.1 ENST00000583534.1 ENST00000457269.4 ENST00000338128.8 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
| chr10_+_60144782 | 1.19 |
ENST00000487519.1 |
TFAM |
transcription factor A, mitochondrial |
| chr10_+_13203543 | 1.18 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr3_-_47823298 | 1.15 |
ENST00000254480.5 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chrX_+_109245863 | 1.14 |
ENST00000372072.3 |
TMEM164 |
transmembrane protein 164 |
| chr21_-_34914394 | 1.14 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr17_-_62097904 | 1.14 |
ENST00000583366.1 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr4_+_39460620 | 1.14 |
ENST00000340169.2 ENST00000261434.3 |
LIAS |
lipoic acid synthetase |
| chr4_+_39460659 | 1.12 |
ENST00000513731.1 |
LIAS |
lipoic acid synthetase |
| chr19_-_18548921 | 1.11 |
ENST00000545187.1 ENST00000578352.1 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
| chr19_+_50887585 | 1.11 |
ENST00000440232.2 ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1 |
polymerase (DNA directed), delta 1, catalytic subunit |
| chrX_-_48755030 | 1.10 |
ENST00000490755.2 ENST00000465150.2 ENST00000495490.2 |
TIMM17B |
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr1_-_200589859 | 1.08 |
ENST00000367350.4 |
KIF14 |
kinesin family member 14 |
| chr6_+_24495067 | 1.08 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr15_+_66797627 | 1.07 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr22_+_21271714 | 1.07 |
ENST00000354336.3 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr17_+_56769924 | 1.07 |
ENST00000461271.1 ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C |
RAD51 paralog C |
| chr4_+_39460689 | 1.07 |
ENST00000381846.1 |
LIAS |
lipoic acid synthetase |
| chr7_-_73153178 | 1.07 |
ENST00000437775.2 ENST00000222800.3 |
ABHD11 |
abhydrolase domain containing 11 |
| chr1_-_246670519 | 1.06 |
ENST00000388985.4 ENST00000490107.1 |
SMYD3 |
SET and MYND domain containing 3 |
| chr10_+_70939983 | 1.03 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr7_-_73153122 | 1.02 |
ENST00000458339.1 |
ABHD11 |
abhydrolase domain containing 11 |
| chr2_-_55496174 | 1.01 |
ENST00000417363.1 ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2 |
mitochondrial translational initiation factor 2 |
| chr6_-_41909191 | 1.01 |
ENST00000512426.1 ENST00000372987.4 |
CCND3 |
cyclin D3 |
| chr12_-_133707021 | 1.00 |
ENST00000537226.1 |
ZNF891 |
zinc finger protein 891 |
| chr16_+_31539197 | 1.00 |
ENST00000564707.1 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr6_+_32121908 | 0.98 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr1_-_52870059 | 0.98 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr8_+_48873479 | 0.97 |
ENST00000262105.2 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr6_+_26183958 | 0.97 |
ENST00000356530.3 |
HIST1H2BE |
histone cluster 1, H2be |
| chrX_-_64754611 | 0.97 |
ENST00000374807.5 ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L |
LAS1-like (S. cerevisiae) |
| chr8_+_48873453 | 0.97 |
ENST00000523944.1 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr1_-_119682812 | 0.95 |
ENST00000537870.1 |
WARS2 |
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr7_+_96747030 | 0.94 |
ENST00000360382.4 |
ACN9 |
ACN9 homolog (S. cerevisiae) |
| chr10_+_22610124 | 0.93 |
ENST00000376663.3 |
BMI1 |
BMI1 polycomb ring finger oncogene |
| chr2_-_55496344 | 0.92 |
ENST00000403721.1 ENST00000263629.4 |
MTIF2 |
mitochondrial translational initiation factor 2 |
| chr11_-_5248294 | 0.92 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr19_+_36486078 | 0.91 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr9_+_132597722 | 0.90 |
ENST00000372429.3 ENST00000315480.4 ENST00000358355.1 |
USP20 |
ubiquitin specific peptidase 20 |
| chr6_-_27114577 | 0.90 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr15_-_23034322 | 0.90 |
ENST00000539711.2 ENST00000560039.1 ENST00000398013.3 ENST00000337451.3 ENST00000359727.4 ENST00000398014.2 |
NIPA2 |
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr18_-_2571437 | 0.90 |
ENST00000574676.1 ENST00000574538.1 ENST00000319888.6 |
METTL4 |
methyltransferase like 4 |
| chr7_+_150065278 | 0.89 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr7_-_73668692 | 0.89 |
ENST00000352131.3 ENST00000055077.3 |
RFC2 |
replication factor C (activator 1) 2, 40kDa |
| chrX_-_11445856 | 0.89 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr5_+_44809027 | 0.88 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr2_+_109335929 | 0.88 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
| chr1_+_91966384 | 0.87 |
ENST00000430031.2 ENST00000234626.6 |
CDC7 |
cell division cycle 7 |
| chr9_-_138853156 | 0.87 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr16_-_103572 | 0.87 |
ENST00000293860.5 |
POLR3K |
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr5_-_68665296 | 0.87 |
ENST00000512152.1 ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9 |
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr1_-_52870104 | 0.86 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr8_-_80942467 | 0.85 |
ENST00000518271.1 ENST00000276585.4 ENST00000521605.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr15_-_75249793 | 0.85 |
ENST00000322177.5 |
RPP25 |
ribonuclease P/MRP 25kDa subunit |
| chr19_+_4402659 | 0.85 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr19_-_41903161 | 0.84 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr12_+_102514019 | 0.83 |
ENST00000537257.1 ENST00000358383.5 ENST00000392911.2 |
PARPBP |
PARP1 binding protein |
| chr1_-_111506562 | 0.82 |
ENST00000485275.2 ENST00000369763.4 |
LRIF1 |
ligand dependent nuclear receptor interacting factor 1 |
| chr12_-_112279694 | 0.81 |
ENST00000443596.1 ENST00000442119.1 |
MAPKAPK5-AS1 |
MAPKAPK5 antisense RNA 1 |
| chr1_-_119683251 | 0.81 |
ENST00000369426.5 ENST00000235521.4 |
WARS2 |
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr14_+_103058948 | 0.81 |
ENST00000262241.6 |
RCOR1 |
REST corepressor 1 |
| chr7_-_73153161 | 0.80 |
ENST00000395147.4 |
ABHD11 |
abhydrolase domain containing 11 |
| chr1_-_15850676 | 0.80 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr16_+_31539183 | 0.80 |
ENST00000302312.4 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr12_+_102513950 | 0.79 |
ENST00000378128.3 ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP |
PARP1 binding protein |
| chrX_-_118986911 | 0.79 |
ENST00000276201.2 ENST00000345865.2 |
UPF3B |
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr7_-_19748640 | 0.78 |
ENST00000222567.5 |
TWISTNB |
TWIST neighbor |
| chrX_+_153991025 | 0.78 |
ENST00000369550.5 |
DKC1 |
dyskeratosis congenita 1, dyskerin |
| chr15_+_78832747 | 0.78 |
ENST00000560217.1 ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr15_+_78833071 | 0.77 |
ENST00000559365.1 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr1_+_150980889 | 0.77 |
ENST00000450884.1 ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE |
prune exopolyphosphatase |
| chrX_-_109561294 | 0.76 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr1_-_229644034 | 0.75 |
ENST00000366678.3 ENST00000261396.3 ENST00000537506.1 |
NUP133 |
nucleoporin 133kDa |
| chr21_-_33651324 | 0.75 |
ENST00000290130.3 |
MIS18A |
MIS18 kinetochore protein A |
| chr10_-_23003460 | 0.74 |
ENST00000376573.4 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr19_+_10527449 | 0.74 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr1_-_15850839 | 0.73 |
ENST00000348549.5 ENST00000546424.1 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr1_+_91966656 | 0.73 |
ENST00000428239.1 ENST00000426137.1 |
CDC7 |
cell division cycle 7 |
| chr1_-_144932316 | 0.73 |
ENST00000313431.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr3_+_142315225 | 0.73 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr17_+_38296576 | 0.73 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr3_+_184080790 | 0.73 |
ENST00000430783.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr4_-_83295103 | 0.72 |
ENST00000313899.7 ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chrX_+_19362011 | 0.72 |
ENST00000379806.5 ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chrX_+_148622138 | 0.71 |
ENST00000450602.2 ENST00000441248.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr2_-_220083671 | 0.70 |
ENST00000439002.2 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr3_+_184081137 | 0.70 |
ENST00000443489.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_+_43333513 | 0.68 |
ENST00000534695.1 ENST00000455725.2 ENST00000531273.1 ENST00000420461.2 ENST00000378852.3 ENST00000534600.1 |
API5 |
apoptosis inhibitor 5 |
| chr1_+_12040238 | 0.68 |
ENST00000444836.1 ENST00000235329.5 |
MFN2 |
mitofusin 2 |
| chr7_+_100472717 | 0.67 |
ENST00000457580.2 ENST00000388793.4 ENST00000432932.1 ENST00000347433.4 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
| chr11_-_78285804 | 0.67 |
ENST00000281038.5 ENST00000529571.1 |
NARS2 |
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr17_-_61850894 | 0.65 |
ENST00000403162.3 ENST00000582252.1 ENST00000225726.5 |
CCDC47 |
coiled-coil domain containing 47 |
| chr4_+_113152978 | 0.65 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_207226313 | 0.65 |
ENST00000367084.1 |
YOD1 |
YOD1 deubiquitinase |
| chr11_+_47600562 | 0.65 |
ENST00000263774.4 ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3 |
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr7_-_140624499 | 0.65 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr1_-_220220000 | 0.65 |
ENST00000366923.3 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr6_+_3068606 | 0.65 |
ENST00000259808.4 |
RIPK1 |
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr12_-_117537240 | 0.64 |
ENST00000392545.4 ENST00000541210.1 ENST00000335209.7 |
TESC |
tescalcin |
| chr8_-_80680078 | 0.64 |
ENST00000337919.5 ENST00000354724.3 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr2_-_220083692 | 0.64 |
ENST00000265316.3 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr8_+_110552337 | 0.63 |
ENST00000337573.5 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr2_-_153573965 | 0.63 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr10_+_35416090 | 0.63 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr14_-_21737551 | 0.62 |
ENST00000554891.1 ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr7_-_128694927 | 0.62 |
ENST00000471166.1 ENST00000265388.5 |
TNPO3 |
transportin 3 |
| chr15_+_40453204 | 0.62 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr14_-_103987679 | 0.62 |
ENST00000553610.1 |
CKB |
creatine kinase, brain |
| chr12_+_120884222 | 0.62 |
ENST00000551765.1 ENST00000229384.5 |
GATC |
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr17_-_40169429 | 0.62 |
ENST00000316603.7 ENST00000588641.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr1_+_150980989 | 0.61 |
ENST00000368935.1 |
PRUNE |
prune exopolyphosphatase |
| chr6_+_34725263 | 0.61 |
ENST00000374018.1 ENST00000374017.3 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr16_-_2301563 | 0.61 |
ENST00000562238.1 ENST00000566379.1 ENST00000301729.4 |
ECI1 |
enoyl-CoA delta isomerase 1 |
| chr7_+_107204389 | 0.60 |
ENST00000265720.3 ENST00000402620.1 |
DUS4L |
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr7_+_120591170 | 0.60 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr5_-_68665084 | 0.60 |
ENST00000509462.1 |
TAF9 |
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr7_-_8301768 | 0.60 |
ENST00000265577.7 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr20_-_5107180 | 0.59 |
ENST00000379160.3 |
PCNA |
proliferating cell nuclear antigen |
| chr11_-_10830463 | 0.59 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_-_109825751 | 0.59 |
ENST00000369907.3 ENST00000438534.2 ENST00000369909.2 ENST00000409138.2 |
PSRC1 |
proline/serine-rich coiled-coil 1 |
| chr3_+_184081213 | 0.59 |
ENST00000429568.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_184081175 | 0.58 |
ENST00000452961.1 ENST00000296223.3 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_-_195808980 | 0.58 |
ENST00000360110.4 |
TFRC |
transferrin receptor |
| chr21_-_33984865 | 0.58 |
ENST00000458138.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr14_-_21737610 | 0.58 |
ENST00000320084.7 ENST00000449098.1 ENST00000336053.6 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr5_-_150080472 | 0.58 |
ENST00000521464.1 ENST00000518917.1 ENST00000447771.2 ENST00000540000.1 ENST00000199814.4 |
RBM22 |
RNA binding motif protein 22 |
| chr7_-_8301869 | 0.57 |
ENST00000402384.3 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr11_+_4116005 | 0.57 |
ENST00000300738.5 |
RRM1 |
ribonucleotide reductase M1 |
| chr11_+_31531291 | 0.56 |
ENST00000350638.5 ENST00000379163.5 ENST00000395934.2 |
ELP4 |
elongator acetyltransferase complex subunit 4 |
| chr10_+_99079008 | 0.56 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr6_-_27100529 | 0.56 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chrX_-_71525742 | 0.56 |
ENST00000450875.1 ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr6_+_27100811 | 0.55 |
ENST00000359193.2 |
HIST1H2AG |
histone cluster 1, H2ag |
| chr10_+_35415719 | 0.55 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr2_-_220094294 | 0.55 |
ENST00000436856.1 ENST00000428226.1 ENST00000409422.1 ENST00000431715.1 ENST00000457841.1 ENST00000439812.1 ENST00000361242.4 ENST00000396761.2 |
ATG9A |
autophagy related 9A |
| chrX_+_148622513 | 0.55 |
ENST00000393985.3 ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr12_+_104359576 | 0.54 |
ENST00000392872.3 ENST00000436021.2 |
TDG |
thymine-DNA glycosylase |
| chr1_-_109825719 | 0.54 |
ENST00000369904.3 ENST00000369903.2 ENST00000429031.1 ENST00000418914.2 ENST00000409267.1 |
PSRC1 |
proline/serine-rich coiled-coil 1 |
| chr17_-_40169161 | 0.54 |
ENST00000589586.2 ENST00000426588.3 ENST00000589576.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr21_-_33984888 | 0.54 |
ENST00000382549.4 ENST00000540881.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr11_+_118955583 | 0.53 |
ENST00000278715.3 ENST00000536813.1 ENST00000537841.1 ENST00000542729.1 ENST00000546302.1 ENST00000442944.2 ENST00000544387.1 ENST00000543090.1 |
HMBS |
hydroxymethylbilane synthase |
| chr15_+_52311398 | 0.53 |
ENST00000261845.5 |
MAPK6 |
mitogen-activated protein kinase 6 |
| chr7_-_8302164 | 0.53 |
ENST00000447326.1 ENST00000406470.2 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr11_-_119252359 | 0.53 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr6_+_108487245 | 0.53 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr1_-_155881156 | 0.53 |
ENST00000539040.1 ENST00000368323.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr1_+_23345943 | 0.53 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr4_-_76439483 | 0.52 |
ENST00000380840.2 ENST00000513257.1 ENST00000507014.1 |
RCHY1 |
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr21_-_33984456 | 0.52 |
ENST00000431216.1 ENST00000553001.1 ENST00000440966.1 |
AP000275.65 C21orf59 |
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr1_-_246729544 | 0.52 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr1_-_200638964 | 0.52 |
ENST00000367348.3 ENST00000447706.2 ENST00000331314.6 |
DDX59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
| chr7_+_39989611 | 0.52 |
ENST00000181839.4 |
CDK13 |
cyclin-dependent kinase 13 |
| chr22_-_42343117 | 0.51 |
ENST00000407253.3 ENST00000215980.5 |
CENPM |
centromere protein M |
| chr20_+_56964253 | 0.51 |
ENST00000395802.3 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_+_104359641 | 0.51 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr1_-_113498943 | 0.51 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr19_-_36231437 | 0.51 |
ENST00000591748.1 |
IGFLR1 |
IGF-like family receptor 1 |
| chr10_+_35416223 | 0.51 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr1_+_23345930 | 0.51 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr12_+_27863706 | 0.51 |
ENST00000081029.3 ENST00000538315.1 ENST00000542791.1 |
MRPS35 |
mitochondrial ribosomal protein S35 |
| chr14_+_75536335 | 0.51 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr21_-_46237883 | 0.51 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr2_+_118572226 | 0.51 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chrX_+_41193407 | 0.50 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr4_+_113152881 | 0.50 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr20_+_31407692 | 0.50 |
ENST00000375571.5 |
MAPRE1 |
microtubule-associated protein, RP/EB family, member 1 |
| chr14_+_75536280 | 0.50 |
ENST00000238686.8 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr5_+_154238149 | 0.50 |
ENST00000519430.1 ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr2_+_88991162 | 0.50 |
ENST00000283646.4 |
RPIA |
ribose 5-phosphate isomerase A |
| chr8_-_124408652 | 0.50 |
ENST00000287394.5 |
ATAD2 |
ATPase family, AAA domain containing 2 |
| chr7_-_128695147 | 0.50 |
ENST00000482320.1 ENST00000393245.1 ENST00000471234.1 |
TNPO3 |
transportin 3 |
| chr1_+_206809113 | 0.50 |
ENST00000441486.1 ENST00000367106.1 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr17_-_40169659 | 0.49 |
ENST00000457167.4 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr9_+_116037922 | 0.49 |
ENST00000374198.4 |
PRPF4 |
pre-mRNA processing factor 4 |
| chr17_+_27071002 | 0.49 |
ENST00000262395.5 ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4 |
TNF receptor-associated factor 4 |
| chr2_-_153573887 | 0.49 |
ENST00000493468.2 ENST00000545856.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr19_-_11639910 | 0.49 |
ENST00000588998.1 ENST00000586149.1 |
ECSIT |
ECSIT signalling integrator |
| chr19_+_17416457 | 0.49 |
ENST00000252602.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr22_-_43539346 | 0.48 |
ENST00000327555.5 ENST00000290429.6 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chrX_+_41192595 | 0.48 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr3_-_186288097 | 0.48 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr9_+_106856831 | 0.48 |
ENST00000303219.8 ENST00000374787.3 |
SMC2 |
structural maintenance of chromosomes 2 |
| chr2_+_61108650 | 0.48 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr7_-_56119156 | 0.47 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 8.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 4.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 7.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 4.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.9 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 1.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.6 | 2.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.6 | 2.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 1.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 1.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 0.8 | GO:0000806 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 2.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 3.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 4.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 1.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 1.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 0.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 0.5 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 2.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.6 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 3.2 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.7 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 9.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 2.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.8 | 3.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.6 | 1.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.5 | 1.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.5 | 1.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 1.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.4 | 1.2 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.4 | 1.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.4 | 1.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.3 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 2.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 1.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.3 | 0.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 | 1.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 0.8 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.1 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.2 | 1.5 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.2 | 1.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 0.7 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.2 | 1.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.2 | 0.9 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 0.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 0.7 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.6 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.2 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 1.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 1.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.7 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.2 | 1.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 0.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 0.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.5 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 0.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.5 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 0.6 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 2.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.1 | 0.4 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.1 | GO:0098840 | intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |
| 0.1 | 1.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.4 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 1.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 3.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 1.0 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 1.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.4 | GO:0019827 | stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.1 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 2.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 1.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 1.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.5 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.4 | GO:0032241 | snRNA export from nucleus(GO:0006408) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 4.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.7 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.6 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.1 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.8 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.7 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.3 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.1 | 1.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 3.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.9 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 1.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 4.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.3 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.3 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.2 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) negative regulation of myeloid dendritic cell activation(GO:0030886) negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.3 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 0.4 | GO:0042262 | DNA protection(GO:0042262) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:2001171 | regulation of ATP biosynthetic process(GO:2001169) positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.1 | 0.3 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 3.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.4 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.4 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.9 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 3.3 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.4 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 3.7 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 1.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.8 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 1.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0070734 | peptidyl-lysine dimethylation(GO:0018027) histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.6 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1904627 | 3'-UTR-mediated mRNA destabilization(GO:0061158) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 2.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 1.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.8 | GO:0043038 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.9 | GO:0030397 | mitotic nuclear envelope disassembly(GO:0007077) membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.0 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0034971 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.5 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.1 | GO:0044550 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 3.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.6 | 2.8 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.5 | 1.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.5 | 2.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 1.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 1.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 1.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.3 | 1.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 0.9 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 4.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.7 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 0.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 0.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 2.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 0.6 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 2.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.9 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 0.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 0.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.2 | 1.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.6 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 1.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.4 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 2.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 1.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 2.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.4 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 3.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.5 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.4 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.7 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 2.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0048273 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.8 | GO:0016787 | hydrolase activity(GO:0016787) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.0 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.9 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |