Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
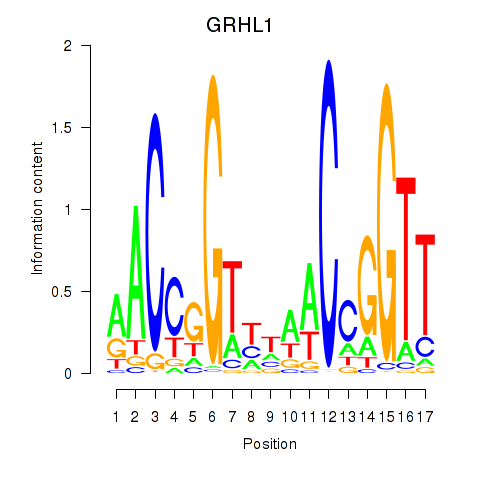
Results for GRHL1
Z-value: 0.94
Transcription factors associated with GRHL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GRHL1
|
ENSG00000134317.13 | GRHL1 |
Activity profile of GRHL1 motif
Sorted Z-values of GRHL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GRHL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_35247859 | 3.67 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr7_-_80551671 | 2.06 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_-_201368707 | 1.91 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr5_+_125758865 | 1.90 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr5_+_125758813 | 1.82 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr1_-_201368653 | 1.80 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr13_+_48807334 | 1.40 |
ENST00000378549.5 |
ITM2B |
integral membrane protein 2B |
| chr19_-_291133 | 1.35 |
ENST00000327790.3 |
PPAP2C |
phosphatidic acid phosphatase type 2C |
| chr13_+_48807288 | 1.33 |
ENST00000378565.5 |
ITM2B |
integral membrane protein 2B |
| chr2_-_85641162 | 1.16 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr20_-_18038521 | 1.12 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr1_-_153113927 | 1.06 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr3_+_101568349 | 1.00 |
ENST00000326151.5 ENST00000326172.5 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chrX_-_119603138 | 0.71 |
ENST00000200639.4 ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2 |
lysosomal-associated membrane protein 2 |
| chr2_-_209118974 | 0.61 |
ENST00000415913.1 ENST00000415282.1 ENST00000446179.1 |
IDH1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr5_-_132299313 | 0.54 |
ENST00000265343.5 |
AFF4 |
AF4/FMR2 family, member 4 |
| chr4_-_99850243 | 0.46 |
ENST00000280892.6 ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr1_+_44440575 | 0.40 |
ENST00000532642.1 ENST00000236067.4 ENST00000471859.2 |
ATP6V0B |
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr16_+_2564254 | 0.39 |
ENST00000565223.1 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr19_+_45312347 | 0.37 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr3_-_172241250 | 0.34 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr7_+_76139741 | 0.33 |
ENST00000334348.3 ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B |
uroplakin 3B |
| chr9_-_77703115 | 0.32 |
ENST00000361092.4 ENST00000376808.4 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr9_+_130565487 | 0.30 |
ENST00000373225.3 ENST00000431857.1 |
FPGS |
folylpolyglutamate synthase |
| chr10_+_81838792 | 0.28 |
ENST00000372273.3 |
TMEM254 |
transmembrane protein 254 |
| chr2_+_113816215 | 0.24 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
| chr19_-_40596767 | 0.23 |
ENST00000599972.1 ENST00000450241.2 ENST00000595687.2 |
ZNF780A |
zinc finger protein 780A |
| chr1_+_155023757 | 0.23 |
ENST00000356955.2 ENST00000449910.2 ENST00000359280.4 ENST00000360674.4 ENST00000368412.3 ENST00000355956.2 ENST00000368410.2 ENST00000271836.6 ENST00000368413.1 ENST00000531455.1 ENST00000447332.3 |
ADAM15 |
ADAM metallopeptidase domain 15 |
| chr9_+_71736177 | 0.21 |
ENST00000606364.1 ENST00000453658.2 |
TJP2 |
tight junction protein 2 |
| chr10_+_105036909 | 0.21 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr12_-_10959892 | 0.21 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr1_+_40723779 | 0.20 |
ENST00000372759.3 |
ZMPSTE24 |
zinc metallopeptidase STE24 |
| chr1_-_24469602 | 0.18 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr10_+_81838411 | 0.16 |
ENST00000372281.3 ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254 |
transmembrane protein 254 |
| chr11_+_15095108 | 0.13 |
ENST00000324229.6 ENST00000533448.1 |
CALCB |
calcitonin-related polypeptide beta |
| chr19_+_12035913 | 0.13 |
ENST00000591944.1 |
ZNF763 |
Uncharacterized protein; Zinc finger protein 763 |
| chr3_+_186383741 | 0.13 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr19_-_17356697 | 0.12 |
ENST00000291442.3 |
NR2F6 |
nuclear receptor subfamily 2, group F, member 6 |
| chr15_+_49170083 | 0.12 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr14_+_23938891 | 0.12 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr12_+_18414446 | 0.11 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr14_-_23540826 | 0.11 |
ENST00000357481.2 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
| chr4_+_110736659 | 0.10 |
ENST00000394631.3 ENST00000226796.6 |
GAR1 |
GAR1 ribonucleoprotein |
| chr7_-_138794081 | 0.10 |
ENST00000464606.1 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
| chr7_-_102184083 | 0.09 |
ENST00000379357.5 |
POLR2J3 |
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr14_-_80677815 | 0.08 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr7_-_102283238 | 0.08 |
ENST00000340457.8 |
UPK3BL |
uroplakin 3B-like |
| chr14_-_80677613 | 0.07 |
ENST00000556811.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr14_-_23540747 | 0.06 |
ENST00000555566.1 ENST00000338631.6 ENST00000557515.1 ENST00000397341.3 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
| chr6_-_31107127 | 0.05 |
ENST00000259845.4 |
PSORS1C2 |
psoriasis susceptibility 1 candidate 2 |
| chr1_-_28384598 | 0.05 |
ENST00000373864.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr10_+_96443204 | 0.04 |
ENST00000339022.5 |
CYP2C18 |
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr7_+_76139925 | 0.03 |
ENST00000394849.1 |
UPK3B |
uroplakin 3B |
| chrX_+_146993449 | 0.03 |
ENST00000218200.8 ENST00000370471.3 ENST00000370477.1 |
FMR1 |
fragile X mental retardation 1 |
| chr19_+_58038683 | 0.03 |
ENST00000240719.3 ENST00000376233.3 ENST00000594943.1 ENST00000602149.1 |
ZNF549 |
zinc finger protein 549 |
| chr11_+_119039414 | 0.03 |
ENST00000409991.1 ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1 |
NLR family member X1 |
| chr1_+_206643787 | 0.02 |
ENST00000367120.3 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr7_+_76139833 | 0.02 |
ENST00000257632.5 |
UPK3B |
uroplakin 3B |
| chr3_-_3152031 | 0.00 |
ENST00000383846.1 ENST00000427088.1 ENST00000446632.2 ENST00000438560.1 |
IL5RA |
interleukin 5 receptor, alpha |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.1 | 2.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 3.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 2.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 1.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.7 | GO:1905146 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 1.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 3.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |