Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for GUAACAG
Z-value: 0.54
miRNA associated with seed GUAACAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-194-5p
|
MIMAT0000460 |
Activity profile of GUAACAG motif
Sorted Z-values of GUAACAG motif
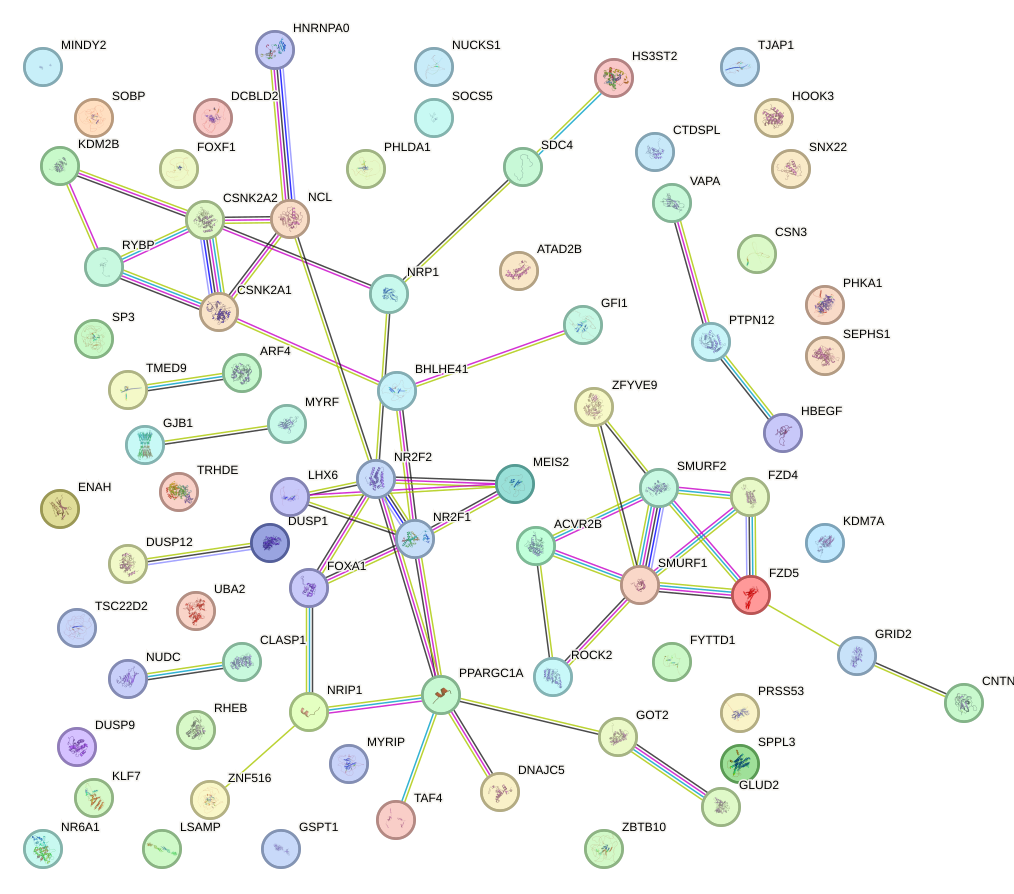
Network of associatons between targets according to the STRING database.
First level regulatory network of GUAACAG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_115375107 | 1.90 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr2_+_102508955 | 0.90 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chr2_-_208634287 | 0.79 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chrX_+_152907913 | 0.68 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr6_+_107811162 | 0.61 |
ENST00000317357.5 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
| chr11_-_86666427 | 0.58 |
ENST00000531380.1 |
FZD4 |
frizzled family receptor 4 |
| chr11_+_61520075 | 0.55 |
ENST00000278836.5 |
MYRF |
myelin regulatory factor |
| chr4_-_23891693 | 0.54 |
ENST00000264867.2 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr11_+_101981169 | 0.54 |
ENST00000526343.1 ENST00000282441.5 ENST00000537274.1 ENST00000345877.2 |
YAP1 |
Yes-associated protein 1 |
| chr20_-_43977055 | 0.52 |
ENST00000372733.3 ENST00000537976.1 |
SDC4 |
syndecan 4 |
| chr12_-_76425368 | 0.51 |
ENST00000602540.1 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
| chr16_+_22825475 | 0.46 |
ENST00000261374.3 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr1_-_225840747 | 0.44 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr16_+_69599861 | 0.40 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr3_+_38495333 | 0.39 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr15_+_96873921 | 0.35 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr10_+_114709999 | 0.32 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_+_70435044 | 0.30 |
ENST00000374029.1 ENST00000374022.3 ENST00000447581.1 |
GJB1 |
gap junction protein, beta 1, 32kDa |
| chr2_-_11484710 | 0.30 |
ENST00000315872.6 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
| chr1_+_82266053 | 0.30 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr3_+_39851094 | 0.26 |
ENST00000302541.6 |
MYRIP |
myosin VIIA and Rab interacting protein |
| chr14_-_38064198 | 0.26 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr12_-_26278030 | 0.26 |
ENST00000242728.4 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
| chr3_+_37903432 | 0.26 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr21_-_16437255 | 0.24 |
ENST00000400199.1 ENST00000400202.1 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr6_+_43445261 | 0.22 |
ENST00000372444.2 ENST00000372445.5 ENST00000436109.2 ENST00000372454.2 ENST00000442878.2 ENST00000259751.1 ENST00000372452.1 ENST00000372449.1 |
TJAP1 |
tight junction associated protein 1 (peripheral) |
| chr1_+_52682052 | 0.21 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr18_+_55102917 | 0.21 |
ENST00000491143.2 |
ONECUT2 |
one cut homeobox 2 |
| chr17_-_57784755 | 0.21 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr3_+_197476621 | 0.20 |
ENST00000241502.4 |
FYTTD1 |
forty-two-three domain containing 1 |
| chr2_-_122407097 | 0.19 |
ENST00000409078.3 |
CLASP1 |
cytoplasmic linker associated protein 1 |
| chr16_-_73082274 | 0.19 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr14_+_55034599 | 0.19 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr8_+_102504651 | 0.19 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chr14_+_39735411 | 0.19 |
ENST00000603904.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr9_-_124989804 | 0.19 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr2_-_174830430 | 0.19 |
ENST00000310015.6 ENST00000455789.2 |
SP3 |
Sp3 transcription factor |
| chr11_+_12695944 | 0.18 |
ENST00000361905.4 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_+_39736299 | 0.16 |
ENST00000341502.5 ENST00000396158.2 ENST00000280083.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr2_+_46926048 | 0.16 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr12_-_121342170 | 0.14 |
ENST00000353487.2 |
SPPL3 |
signal peptide peptidase like 3 |
| chr10_-_13390270 | 0.14 |
ENST00000378614.4 ENST00000545675.1 ENST00000327347.5 |
SEPHS1 |
selenophosphate synthetase 1 |
| chr5_-_172198190 | 0.13 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr3_+_9439400 | 0.12 |
ENST00000450326.1 ENST00000402198.1 ENST00000402466.1 |
SETD5 |
SET domain containing 5 |
| chr16_-_12009735 | 0.12 |
ENST00000439887.2 ENST00000434724.2 |
GSPT1 |
G1 to S phase transition 1 |
| chr9_-_127533519 | 0.12 |
ENST00000487099.2 ENST00000344523.4 ENST00000373584.3 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chrX_-_71933888 | 0.12 |
ENST00000373542.4 ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
| chr20_-_524455 | 0.11 |
ENST00000349736.5 ENST00000217244.3 |
CSNK2A1 |
casein kinase 2, alpha 1 polypeptide |
| chr7_-_151217001 | 0.11 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr7_-_98741642 | 0.11 |
ENST00000361368.2 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr14_-_21905395 | 0.10 |
ENST00000430710.3 ENST00000553283.1 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
| chr5_-_115910630 | 0.10 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_-_98620500 | 0.10 |
ENST00000326840.6 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
| chr16_-_31100284 | 0.10 |
ENST00000280606.6 |
PRSS53 |
protease, serine, 53 |
| chr8_+_81397876 | 0.10 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr1_+_27248203 | 0.10 |
ENST00000321265.5 |
NUDC |
nudC nuclear distribution protein |
| chr20_-_60640866 | 0.09 |
ENST00000252996.4 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr5_+_177019159 | 0.09 |
ENST00000332598.6 |
TMED9 |
transmembrane emp24 protein transport domain containing 9 |
| chr16_-_58768177 | 0.09 |
ENST00000434819.2 ENST00000245206.5 |
GOT2 |
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr18_-_74207146 | 0.09 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr18_+_9913977 | 0.09 |
ENST00000400000.2 ENST00000340541.4 |
VAPA |
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr8_+_42752053 | 0.09 |
ENST00000307602.4 |
HOOK3 |
hook microtubule-tethering protein 3 |
| chr10_-_33623564 | 0.09 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr12_-_88974236 | 0.08 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chr15_+_64443905 | 0.08 |
ENST00000325881.4 |
SNX22 |
sorting nexin 22 |
| chr5_-_98262240 | 0.08 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr1_-_205719295 | 0.08 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chrX_-_41782249 | 0.08 |
ENST00000442742.2 ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr2_-_232329186 | 0.08 |
ENST00000322723.4 |
NCL |
nucleolin |
| chr15_+_59063478 | 0.08 |
ENST00000559228.1 ENST00000450403.2 |
FAM63B |
family with sequence similarity 63, member B |
| chr3_+_150126101 | 0.08 |
ENST00000361875.3 ENST00000361136.2 |
TSC22D2 |
TSC22 domain family, member 2 |
| chr2_-_208030647 | 0.07 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr7_-_139876812 | 0.07 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr17_-_4167142 | 0.07 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr5_-_137090028 | 0.07 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr7_+_77166592 | 0.07 |
ENST00000248594.6 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr12_+_72666407 | 0.07 |
ENST00000261180.4 |
TRHDE |
thyrotropin-releasing hormone degrading enzyme |
| chr3_-_116164306 | 0.07 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr5_-_139726181 | 0.07 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr2_-_24149977 | 0.07 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr1_-_92949505 | 0.06 |
ENST00000370332.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr15_-_37390482 | 0.06 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr12_-_122018859 | 0.06 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr3_-_72496035 | 0.06 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr3_-_57583130 | 0.06 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr16_+_86544113 | 0.06 |
ENST00000262426.4 |
FOXF1 |
forkhead box F1 |
| chrX_+_120181457 | 0.05 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr17_-_62658186 | 0.05 |
ENST00000262435.9 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
| chr7_+_145813453 | 0.05 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr19_+_34919257 | 0.05 |
ENST00000246548.4 ENST00000590048.2 |
UBA2 |
ubiquitin-like modifier activating enzyme 2 |
| chr19_-_2151523 | 0.05 |
ENST00000350812.6 ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1 |
adaptor-related protein complex 3, delta 1 subunit |
| chr15_+_62853562 | 0.05 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr17_-_80231557 | 0.04 |
ENST00000392334.2 ENST00000314028.6 |
CSNK1D |
casein kinase 1, delta |
| chr6_-_154831779 | 0.04 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr17_-_73179046 | 0.04 |
ENST00000314523.7 ENST00000420826.2 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr9_-_91793675 | 0.04 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr3_-_149688896 | 0.04 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr19_-_14606900 | 0.04 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr2_+_210444142 | 0.04 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr15_+_39873268 | 0.03 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr4_-_57301748 | 0.03 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr17_+_40985407 | 0.03 |
ENST00000586114.1 ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3 |
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr17_-_33416231 | 0.03 |
ENST00000584655.1 ENST00000447669.2 ENST00000315249.7 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr7_+_77325738 | 0.03 |
ENST00000334955.8 |
RSBN1L |
round spermatid basic protein 1-like |
| chr3_-_58419537 | 0.03 |
ENST00000474765.1 ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB |
pyruvate dehydrogenase (lipoamide) beta |
| chr3_+_14166440 | 0.03 |
ENST00000306077.4 |
TMEM43 |
transmembrane protein 43 |
| chr8_-_116681221 | 0.03 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr21_-_34960948 | 0.02 |
ENST00000453626.1 ENST00000303113.6 ENST00000432378.1 ENST00000303071.5 |
DONSON |
downstream neighbor of SON |
| chr17_+_37618257 | 0.02 |
ENST00000447079.4 |
CDK12 |
cyclin-dependent kinase 12 |
| chr3_+_37493610 | 0.02 |
ENST00000264741.5 |
ITGA9 |
integrin, alpha 9 |
| chr14_-_77843390 | 0.02 |
ENST00000216468.7 |
TMED8 |
transmembrane emp24 protein transport domain containing 8 |
| chr1_-_52456352 | 0.02 |
ENST00000371655.3 |
RAB3B |
RAB3B, member RAS oncogene family |
| chr4_-_66536057 | 0.02 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr13_+_42622781 | 0.02 |
ENST00000337343.4 ENST00000261491.5 ENST00000379274.2 |
DGKH |
diacylglycerol kinase, eta |
| chr12_+_123868320 | 0.02 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr1_-_93645818 | 0.02 |
ENST00000370280.1 ENST00000479918.1 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
| chr10_-_72648541 | 0.02 |
ENST00000299299.3 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr17_+_61699766 | 0.02 |
ENST00000579585.1 ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
| chr3_-_133614597 | 0.02 |
ENST00000285208.4 ENST00000460865.3 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr22_+_40390930 | 0.02 |
ENST00000333407.6 |
FAM83F |
family with sequence similarity 83, member F |
| chr6_+_163835669 | 0.01 |
ENST00000453779.2 ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI |
QKI, KH domain containing, RNA binding |
| chr1_+_2985760 | 0.01 |
ENST00000378391.2 ENST00000514189.1 ENST00000270722.5 |
PRDM16 |
PR domain containing 16 |
| chr10_+_105726862 | 0.01 |
ENST00000335753.4 ENST00000369755.3 |
SLK |
STE20-like kinase |
| chr9_+_108006880 | 0.01 |
ENST00000374723.1 ENST00000374720.3 ENST00000374724.1 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
| chr17_+_48796905 | 0.01 |
ENST00000505658.1 ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr7_+_138916231 | 0.01 |
ENST00000473989.3 ENST00000288561.8 |
UBN2 |
ubinuclein 2 |
| chr14_-_93799360 | 0.01 |
ENST00000334746.5 ENST00000554565.1 ENST00000298896.3 |
BTBD7 |
BTB (POZ) domain containing 7 |
| chr1_-_11120057 | 0.01 |
ENST00000376957.2 |
SRM |
spermidine synthase |
| chr9_+_129677039 | 0.01 |
ENST00000259351.5 ENST00000424082.2 ENST00000394022.3 ENST00000394011.3 ENST00000319107.4 |
RALGPS1 |
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_-_153958805 | 0.00 |
ENST00000368575.3 |
RAB13 |
RAB13, member RAS oncogene family |
| chr8_-_74791051 | 0.00 |
ENST00000453587.2 ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
| chr2_+_85198216 | 0.00 |
ENST00000456682.1 ENST00000409785.4 |
KCMF1 |
potassium channel modulatory factor 1 |
| chr3_+_196466710 | 0.00 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr17_+_35294075 | 0.00 |
ENST00000254457.5 |
LHX1 |
LIM homeobox 1 |
| chr10_-_75634260 | 0.00 |
ENST00000372765.1 ENST00000351293.3 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 0.8 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 0.5 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.3 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.4 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |