Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
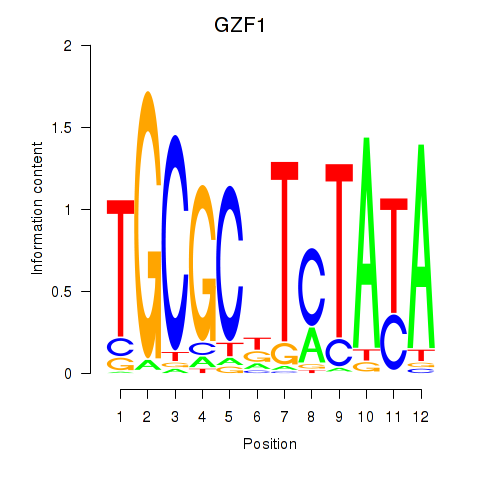
Results for GZF1
Z-value: 0.79
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GZF1 |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_52535712 | 3.12 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr3_-_87040233 | 2.12 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr3_-_114035026 | 1.78 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr18_-_53303123 | 1.68 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr1_+_26606608 | 1.19 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr1_-_144995002 | 1.08 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chrX_+_54835493 | 1.02 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr1_-_144994909 | 0.84 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_-_144995074 | 0.83 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr14_-_92413353 | 0.78 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr10_-_126849588 | 0.76 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr10_-_126849068 | 0.75 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr14_-_92413727 | 0.70 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr3_+_30647994 | 0.69 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr5_+_92919043 | 0.58 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_30648066 | 0.57 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr14_+_101292445 | 0.53 |
ENST00000429159.2 ENST00000520714.1 ENST00000522771.2 ENST00000424076.3 ENST00000423456.1 ENST00000521404.1 ENST00000556736.1 ENST00000451743.2 ENST00000398518.2 ENST00000554639.1 ENST00000452120.2 ENST00000519709.1 ENST00000412736.2 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr20_+_42086525 | 0.44 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr9_+_96338647 | 0.41 |
ENST00000359246.4 |
PHF2 |
PHD finger protein 2 |
| chr1_-_144994840 | 0.41 |
ENST00000369351.3 ENST00000369349.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_-_226374373 | 0.39 |
ENST00000366812.5 |
ACBD3 |
acyl-CoA binding domain containing 3 |
| chr7_-_27183263 | 0.33 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chrX_+_102883620 | 0.32 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr12_+_107168418 | 0.30 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr1_-_32801825 | 0.28 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr9_+_96338860 | 0.23 |
ENST00000375376.4 |
PHF2 |
PHD finger protein 2 |
| chr12_+_104458235 | 0.23 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr9_-_14308004 | 0.21 |
ENST00000493697.1 |
NFIB |
nuclear factor I/B |
| chr9_+_131133598 | 0.16 |
ENST00000372853.4 ENST00000452446.1 ENST00000372850.1 ENST00000372847.1 |
URM1 |
ubiquitin related modifier 1 |
| chr12_-_76478417 | 0.13 |
ENST00000552342.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr12_-_76478386 | 0.13 |
ENST00000535020.2 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr14_-_21979428 | 0.11 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr10_+_89264625 | 0.10 |
ENST00000371996.4 ENST00000371994.4 |
MINPP1 |
multiple inositol-polyphosphate phosphatase 1 |
| chr2_-_150444300 | 0.09 |
ENST00000303319.5 |
MMADHC |
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr12_-_76478446 | 0.08 |
ENST00000393263.3 ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr14_+_22293618 | 0.07 |
ENST00000390432.2 |
TRAV10 |
T cell receptor alpha variable 10 |
| chr2_-_150444116 | 0.07 |
ENST00000428879.1 ENST00000422782.2 |
MMADHC |
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr7_+_102004322 | 0.06 |
ENST00000496391.1 |
PRKRIP1 |
PRKR interacting protein 1 (IL11 inducible) |
| chr7_-_105029812 | 0.04 |
ENST00000482897.1 |
SRPK2 |
SRSF protein kinase 2 |
| chr6_+_30524663 | 0.02 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr6_+_30525051 | 0.01 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.6 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 3.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.2 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 1.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |