Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HBP1
Z-value: 1.33
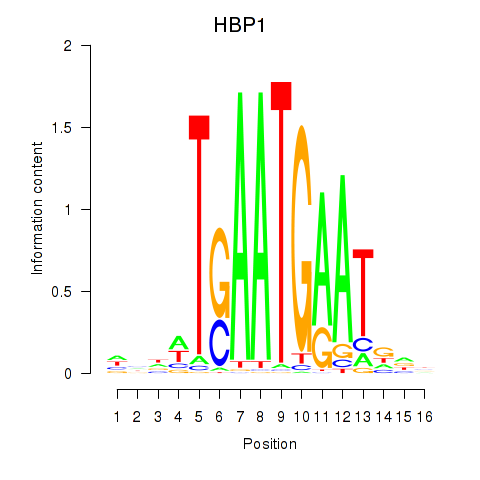
Transcription factors associated with HBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HBP1
|
ENSG00000105856.9 | HBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HBP1 | hg19_v2_chr7_+_106809406_106809460 | -0.50 | 4.9e-02 | Click! |
Activity profile of HBP1 motif
Sorted Z-values of HBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_6690723 | 4.34 |
ENST00000601008.1 |
C3 |
complement component 3 |
| chr19_-_10697895 | 3.35 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr12_-_89746173 | 2.98 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr2_-_113594279 | 2.51 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr12_-_53320245 | 2.51 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr11_+_66824276 | 2.00 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr2_+_173292390 | 1.73 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292280 | 1.72 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292301 | 1.51 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chrX_+_150151824 | 1.48 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr20_-_56286479 | 1.38 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr2_-_161056762 | 1.36 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr11_+_66824346 | 1.32 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr5_-_78809950 | 1.31 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chrX_+_150151752 | 1.30 |
ENST00000325307.7 |
HMGB3 |
high mobility group box 3 |
| chr2_-_161056802 | 1.29 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr11_-_66725837 | 1.23 |
ENST00000393958.2 ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC |
pyruvate carboxylase |
| chr1_-_24126892 | 1.20 |
ENST00000374497.3 ENST00000425913.1 |
GALE |
UDP-galactose-4-epimerase |
| chr12_+_100867486 | 1.12 |
ENST00000548884.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr4_+_100495864 | 1.04 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr1_-_17338267 | 0.94 |
ENST00000326735.8 |
ATP13A2 |
ATPase type 13A2 |
| chr3_+_46616017 | 0.93 |
ENST00000542931.1 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr20_+_25176318 | 0.92 |
ENST00000354989.5 ENST00000360031.2 ENST00000376652.4 ENST00000439162.1 ENST00000433417.1 ENST00000417467.1 ENST00000433259.2 ENST00000427553.1 ENST00000435520.1 ENST00000418890.1 |
ENTPD6 |
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr3_-_52090461 | 0.85 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr16_-_82203780 | 0.85 |
ENST00000563504.1 ENST00000569021.1 ENST00000258169.4 |
MPHOSPH6 |
M-phase phosphoprotein 6 |
| chr12_+_100867694 | 0.85 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr18_-_19283649 | 0.75 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr8_-_8751068 | 0.73 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr1_+_65613340 | 0.71 |
ENST00000546702.1 |
AK4 |
adenylate kinase 4 |
| chr1_+_13516066 | 0.70 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr12_-_58027138 | 0.68 |
ENST00000341156.4 |
B4GALNT1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr1_+_13736907 | 0.68 |
ENST00000316412.5 |
PRAMEF20 |
PRAME family member 20 |
| chr1_+_65730385 | 0.65 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr12_+_57984965 | 0.65 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr19_-_42806723 | 0.62 |
ENST00000262890.3 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr20_-_33872548 | 0.61 |
ENST00000374443.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr11_+_22689648 | 0.61 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr19_-_18508396 | 0.60 |
ENST00000595840.1 ENST00000339007.3 |
LRRC25 |
leucine rich repeat containing 25 |
| chr22_-_20368028 | 0.60 |
ENST00000404912.1 |
GGTLC3 |
gamma-glutamyltransferase light chain 3 |
| chr2_+_231902193 | 0.59 |
ENST00000373640.4 |
C2orf72 |
chromosome 2 open reading frame 72 |
| chr8_-_130951940 | 0.59 |
ENST00000522250.1 ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr17_-_27405875 | 0.57 |
ENST00000359450.6 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr11_-_18343669 | 0.57 |
ENST00000396253.3 ENST00000349215.3 ENST00000438420.2 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
| chr3_-_197026152 | 0.57 |
ENST00000419354.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr22_-_20367797 | 0.56 |
ENST00000424787.2 |
GGTLC3 |
gamma-glutamyltransferase light chain 3 |
| chr3_+_111805182 | 0.56 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr14_-_94759361 | 0.55 |
ENST00000393096.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759408 | 0.54 |
ENST00000554723.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr15_-_56535464 | 0.50 |
ENST00000559447.2 ENST00000422057.1 ENST00000317318.6 ENST00000423270.1 |
RFX7 |
regulatory factor X, 7 |
| chr5_+_149737202 | 0.50 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr22_+_22988816 | 0.50 |
ENST00000480559.1 ENST00000448514.1 |
GGTLC2 |
gamma-glutamyltransferase light chain 2 |
| chr14_-_94759595 | 0.48 |
ENST00000261994.4 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr19_-_42806444 | 0.48 |
ENST00000594989.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr19_+_1040096 | 0.47 |
ENST00000263094.6 ENST00000524850.1 |
ABCA7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr2_+_74425689 | 0.47 |
ENST00000394053.2 ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr20_-_33543546 | 0.47 |
ENST00000216951.2 |
GSS |
glutathione synthetase |
| chr3_+_184032419 | 0.47 |
ENST00000352767.3 ENST00000427141.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr7_+_39125365 | 0.46 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr1_+_65613217 | 0.45 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr3_-_30936153 | 0.45 |
ENST00000454381.3 ENST00000282538.5 |
GADL1 |
glutamate decarboxylase-like 1 |
| chr8_-_11725549 | 0.45 |
ENST00000505496.2 ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB |
cathepsin B |
| chr11_-_65667997 | 0.44 |
ENST00000312562.2 ENST00000534222.1 |
FOSL1 |
FOS-like antigen 1 |
| chr1_+_44435646 | 0.44 |
ENST00000255108.3 ENST00000412950.2 ENST00000396758.2 |
DPH2 |
DPH2 homolog (S. cerevisiae) |
| chr18_+_18943554 | 0.43 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr7_+_95115210 | 0.43 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr11_-_65667884 | 0.43 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr6_+_30297306 | 0.42 |
ENST00000420746.1 ENST00000513556.1 |
TRIM39 TRIM39-RPP21 |
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr7_+_29874341 | 0.42 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chrX_+_100075368 | 0.42 |
ENST00000415585.2 ENST00000372972.2 ENST00000413437.1 |
CSTF2 |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
| chr20_-_33872518 | 0.42 |
ENST00000374436.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr19_-_42806919 | 0.42 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr22_-_30956746 | 0.39 |
ENST00000437282.1 ENST00000447224.1 ENST00000427899.1 ENST00000406955.1 ENST00000452827.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr1_+_158149737 | 0.39 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr7_-_56101826 | 0.39 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr20_-_23967432 | 0.37 |
ENST00000286890.4 ENST00000278765.4 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
| chr17_+_33895090 | 0.37 |
ENST00000592381.1 |
RP11-1094M14.11 |
RP11-1094M14.11 |
| chr10_-_75457554 | 0.37 |
ENST00000374094.4 ENST00000443782.2 |
AGAP5 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 |
| chr16_+_57769635 | 0.35 |
ENST00000379661.3 ENST00000562592.1 ENST00000566726.1 |
KATNB1 |
katanin p80 (WD repeat containing) subunit B 1 |
| chr3_+_51575596 | 0.35 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr3_+_184032283 | 0.35 |
ENST00000346169.2 ENST00000414031.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_+_44626446 | 0.35 |
ENST00000441021.1 ENST00000322734.2 |
ZNF660 |
zinc finger protein 660 |
| chr7_+_18535786 | 0.35 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr3_-_197025447 | 0.35 |
ENST00000346964.2 ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr12_-_122018859 | 0.35 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr3_+_184032313 | 0.33 |
ENST00000392537.2 ENST00000444134.1 ENST00000450424.1 ENST00000421110.1 ENST00000382330.3 ENST00000426123.1 ENST00000350481.5 ENST00000455679.1 ENST00000440448.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr16_-_20753114 | 0.30 |
ENST00000396083.2 |
THUMPD1 |
THUMP domain containing 1 |
| chr19_-_52034971 | 0.29 |
ENST00000346477.3 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr6_-_133035185 | 0.29 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr1_+_23345930 | 0.28 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr1_+_23345943 | 0.28 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr8_-_86290333 | 0.28 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr12_-_71533055 | 0.28 |
ENST00000552128.1 |
TSPAN8 |
tetraspanin 8 |
| chr19_-_45927622 | 0.27 |
ENST00000300853.3 ENST00000589165.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr2_-_239197201 | 0.27 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr17_-_42143963 | 0.27 |
ENST00000585388.1 ENST00000293406.3 |
LSM12 |
LSM12 homolog (S. cerevisiae) |
| chr19_-_52035044 | 0.27 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr11_+_122709200 | 0.27 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr4_-_186696425 | 0.26 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr17_-_42327236 | 0.26 |
ENST00000399246.2 |
AC003102.1 |
AC003102.1 |
| chr6_-_30640811 | 0.26 |
ENST00000376442.3 |
DHX16 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr10_+_13203543 | 0.25 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr19_+_18794470 | 0.24 |
ENST00000321949.8 ENST00000338797.6 |
CRTC1 |
CREB regulated transcription coactivator 1 |
| chr19_-_6333614 | 0.24 |
ENST00000301452.4 |
ACER1 |
alkaline ceramidase 1 |
| chr17_+_75369167 | 0.24 |
ENST00000423034.2 |
SEPT9 |
septin 9 |
| chr21_+_45138941 | 0.24 |
ENST00000398081.1 ENST00000468090.1 ENST00000291565.4 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr6_+_42531798 | 0.23 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr9_+_78505554 | 0.23 |
ENST00000545128.1 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
| chr12_+_123868320 | 0.23 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr5_-_147211226 | 0.22 |
ENST00000296695.5 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr6_+_10521574 | 0.22 |
ENST00000495262.1 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr5_+_150040403 | 0.22 |
ENST00000517768.1 ENST00000297130.4 |
MYOZ3 |
myozenin 3 |
| chr5_+_140514782 | 0.22 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr2_+_235887329 | 0.22 |
ENST00000409212.1 ENST00000344528.4 ENST00000444916.1 |
SH3BP4 |
SH3-domain binding protein 4 |
| chr22_+_25615489 | 0.22 |
ENST00000398215.2 |
CRYBB2 |
crystallin, beta B2 |
| chr11_-_108369101 | 0.21 |
ENST00000323468.5 |
KDELC2 |
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr2_-_40739501 | 0.21 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr12_-_89919965 | 0.20 |
ENST00000548729.1 |
POC1B-GALNT4 |
POC1B-GALNT4 readthrough |
| chr7_+_99070464 | 0.20 |
ENST00000331410.5 ENST00000483089.1 ENST00000448667.1 ENST00000493485.1 |
ZNF789 |
zinc finger protein 789 |
| chr14_+_102430855 | 0.20 |
ENST00000360184.4 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
| chr19_-_3500635 | 0.20 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr9_+_127624387 | 0.19 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr6_-_8102279 | 0.19 |
ENST00000488226.2 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
| chr1_-_53163992 | 0.19 |
ENST00000371538.3 |
SELRC1 |
cytochrome c oxidase assembly factor 7 |
| chr2_-_30144432 | 0.18 |
ENST00000389048.3 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
| chr1_+_12976450 | 0.17 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr11_-_3818688 | 0.17 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr19_+_49055332 | 0.17 |
ENST00000201586.2 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
| chr2_+_65216462 | 0.17 |
ENST00000234256.3 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_+_207630081 | 0.17 |
ENST00000236980.6 ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2 |
FAST kinase domains 2 |
| chr7_-_83278322 | 0.16 |
ENST00000307792.3 |
SEMA3E |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr5_-_140070897 | 0.16 |
ENST00000448240.1 ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS |
histidyl-tRNA synthetase |
| chr17_-_1588101 | 0.16 |
ENST00000577001.1 ENST00000572621.1 ENST00000304992.6 |
PRPF8 |
pre-mRNA processing factor 8 |
| chr11_-_47574664 | 0.16 |
ENST00000310513.5 ENST00000531165.1 |
CELF1 |
CUGBP, Elav-like family member 1 |
| chrY_+_4868267 | 0.16 |
ENST00000333703.4 |
PCDH11Y |
protocadherin 11 Y-linked |
| chr3_-_54962100 | 0.16 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr2_-_20850813 | 0.16 |
ENST00000402541.1 ENST00000406618.3 ENST00000304031.3 |
HS1BP3 |
HCLS1 binding protein 3 |
| chr15_-_86338134 | 0.16 |
ENST00000337975.5 |
KLHL25 |
kelch-like family member 25 |
| chr11_+_18343800 | 0.16 |
ENST00000453096.2 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr1_+_52082751 | 0.16 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr20_+_45523227 | 0.15 |
ENST00000327619.5 ENST00000357410.3 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
| chr1_+_46972668 | 0.15 |
ENST00000371956.4 ENST00000360032.3 |
DMBX1 |
diencephalon/mesencephalon homeobox 1 |
| chr11_-_62457371 | 0.15 |
ENST00000317449.4 |
LRRN4CL |
LRRN4 C-terminal like |
| chr1_-_20306909 | 0.14 |
ENST00000375111.3 ENST00000400520.3 |
PLA2G2A |
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr16_+_31366455 | 0.14 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_-_151319710 | 0.14 |
ENST00000290524.4 ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr9_+_78505581 | 0.14 |
ENST00000376767.3 ENST00000376752.4 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
| chr2_-_27886460 | 0.13 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr9_+_131217459 | 0.13 |
ENST00000497812.2 ENST00000393533.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr11_+_6947720 | 0.12 |
ENST00000414517.2 |
ZNF215 |
zinc finger protein 215 |
| chr2_-_239197238 | 0.12 |
ENST00000254657.3 |
PER2 |
period circadian clock 2 |
| chr12_-_18243075 | 0.12 |
ENST00000536890.1 |
RERGL |
RERG/RAS-like |
| chr22_+_24236191 | 0.12 |
ENST00000215754.7 |
MIF |
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr8_+_23104130 | 0.11 |
ENST00000313219.7 ENST00000519984.1 |
CHMP7 |
charged multivesicular body protein 7 |
| chr12_-_89920030 | 0.11 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr11_-_57158109 | 0.11 |
ENST00000525955.1 ENST00000533605.1 ENST00000311862.5 |
PRG2 |
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chr17_-_36904437 | 0.10 |
ENST00000585100.1 ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr19_-_38806560 | 0.10 |
ENST00000591755.1 ENST00000337679.8 ENST00000339413.6 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr10_+_81838411 | 0.10 |
ENST00000372281.3 ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254 |
transmembrane protein 254 |
| chr1_-_24127256 | 0.10 |
ENST00000418277.1 |
GALE |
UDP-galactose-4-epimerase |
| chr15_+_75074385 | 0.10 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr8_-_82359662 | 0.09 |
ENST00000519260.1 ENST00000256103.2 |
PMP2 |
peripheral myelin protein 2 |
| chr1_+_92632542 | 0.09 |
ENST00000409154.4 ENST00000370378.4 |
KIAA1107 |
KIAA1107 |
| chr19_+_19144384 | 0.09 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr7_-_99277610 | 0.09 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr17_-_41856305 | 0.09 |
ENST00000397937.2 ENST00000226004.3 |
DUSP3 |
dual specificity phosphatase 3 |
| chrX_+_2746818 | 0.08 |
ENST00000398806.3 |
GYG2 |
glycogenin 2 |
| chr19_-_19144243 | 0.08 |
ENST00000594445.1 ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2 |
SURP and G patch domain containing 2 |
| chr6_+_149539767 | 0.08 |
ENST00000606202.1 ENST00000536230.1 ENST00000445901.1 |
TAB2 RP1-111D6.3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr2_-_73053126 | 0.08 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr2_-_220264703 | 0.08 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr7_+_65540780 | 0.07 |
ENST00000304874.9 |
ASL |
argininosuccinate lyase |
| chr2_+_27886330 | 0.07 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_-_91398796 | 0.07 |
ENST00000261172.3 ENST00000551767.1 |
EPYC |
epiphycan |
| chr19_-_10305752 | 0.07 |
ENST00000540357.1 ENST00000359526.4 ENST00000340748.4 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
| chr3_+_184279566 | 0.07 |
ENST00000330394.2 |
EPHB3 |
EPH receptor B3 |
| chr8_-_16035454 | 0.07 |
ENST00000355282.2 |
MSR1 |
macrophage scavenger receptor 1 |
| chr11_+_6947647 | 0.06 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr5_-_22853429 | 0.06 |
ENST00000504376.2 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
| chr7_+_65540853 | 0.06 |
ENST00000380839.4 ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL |
argininosuccinate lyase |
| chr4_+_6202448 | 0.06 |
ENST00000508601.1 |
RP11-586D19.1 |
RP11-586D19.1 |
| chr6_+_29555683 | 0.05 |
ENST00000383640.2 |
OR2H2 |
olfactory receptor, family 2, subfamily H, member 2 |
| chr5_+_96038476 | 0.05 |
ENST00000511049.1 ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST |
calpastatin |
| chr1_-_13117736 | 0.05 |
ENST00000376192.5 ENST00000376182.1 |
PRAMEF6 |
PRAME family member 6 |
| chr19_+_3506261 | 0.05 |
ENST00000441788.2 |
FZR1 |
fizzy/cell division cycle 20 related 1 (Drosophila) |
| chr5_+_161494521 | 0.05 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_-_32339671 | 0.04 |
ENST00000442822.2 ENST00000375015.4 ENST00000533191.1 |
C6orf10 |
chromosome 6 open reading frame 10 |
| chr11_-_89653576 | 0.04 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chrX_+_2746850 | 0.04 |
ENST00000381163.3 ENST00000338623.5 ENST00000542787.1 |
GYG2 |
glycogenin 2 |
| chr1_-_13115578 | 0.04 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr19_+_34855925 | 0.03 |
ENST00000590375.1 ENST00000356487.5 |
GPI |
glucose-6-phosphate isomerase |
| chr12_-_18243119 | 0.03 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr8_+_110098850 | 0.03 |
ENST00000518632.1 |
TRHR |
thyrotropin-releasing hormone receptor |
| chr16_+_31366536 | 0.02 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr22_-_22337146 | 0.02 |
ENST00000398793.2 |
TOP3B |
topoisomerase (DNA) III beta |
| chr3_-_123710199 | 0.02 |
ENST00000184183.4 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr12_-_110271178 | 0.02 |
ENST00000261740.2 ENST00000392719.2 ENST00000346520.2 |
TRPV4 |
transient receptor potential cation channel, subfamily V, member 4 |
| chr17_+_49230897 | 0.02 |
ENST00000393196.3 ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1 NME1-NME2 NME2 |
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr5_+_161274940 | 0.02 |
ENST00000393943.4 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr5_+_161274685 | 0.01 |
ENST00000428797.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr13_-_46626847 | 0.01 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 3.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 4.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 3.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.9 | 2.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 0.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 3.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0099572 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) postsynaptic specialization(GO:0099572) |
| 0.0 | 4.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.6 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 4.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 1.3 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 5.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 1.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 3.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.6 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.2 | 0.9 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 0.5 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.5 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 0.9 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 0.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.2 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 2.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.6 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.1 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.4 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 2.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 6.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.4 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.8 | 2.5 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.7 | 3.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 2.0 | GO:2001250 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 1.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 1.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 5.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.3 | 0.9 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.3 | 2.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 1.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.2 | 0.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 2.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.5 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.2 | 0.5 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 2.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 1.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 1.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.1 | 1.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.6 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 1.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.4 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.2 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 3.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 3.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.9 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.9 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.4 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 2.3 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.0 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.5 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |