Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HDX
Z-value: 0.73
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | HDX |
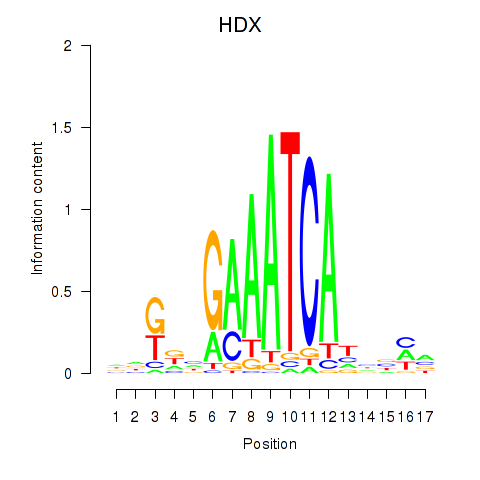
Activity profile of HDX motif
Sorted Z-values of HDX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113594279 | 3.57 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr1_-_153363452 | 3.36 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr1_-_209825674 | 2.97 |
ENST00000367030.3 ENST00000356082.4 |
LAMB3 |
laminin, beta 3 |
| chr18_+_29027696 | 1.97 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr12_+_54422142 | 1.18 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr2_+_113735575 | 0.37 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr5_+_140165876 | 0.33 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr5_-_35230771 | 0.26 |
ENST00000342362.5 |
PRLR |
prolactin receptor |
| chr5_-_41213607 | 0.22 |
ENST00000337836.5 ENST00000433294.1 |
C6 |
complement component 6 |
| chr22_+_40440804 | 0.20 |
ENST00000441751.1 ENST00000301923.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr5_-_35230649 | 0.17 |
ENST00000382002.5 |
PRLR |
prolactin receptor |
| chr11_+_9685604 | 0.16 |
ENST00000447399.2 ENST00000318950.6 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
| chr1_+_117602925 | 0.14 |
ENST00000369466.4 |
TTF2 |
transcription termination factor, RNA polymerase II |
| chr19_+_36602104 | 0.13 |
ENST00000585332.1 ENST00000262637.4 |
OVOL3 |
ovo-like zinc finger 3 |
| chr5_+_140625147 | 0.12 |
ENST00000231173.3 |
PCDHB15 |
protocadherin beta 15 |
| chr1_-_48866517 | 0.11 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr1_-_159684371 | 0.10 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr1_+_84630053 | 0.09 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr12_+_10460549 | 0.09 |
ENST00000543420.1 ENST00000543777.1 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr8_+_24151620 | 0.08 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr11_-_66675371 | 0.05 |
ENST00000393955.2 |
PC |
pyruvate carboxylase |
| chr3_+_54157480 | 0.05 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr5_-_135290705 | 0.04 |
ENST00000274507.1 |
LECT2 |
leukocyte cell-derived chemotaxin 2 |
| chr8_+_110098850 | 0.04 |
ENST00000518632.1 |
TRHR |
thyrotropin-releasing hormone receptor |
| chr12_+_10460417 | 0.03 |
ENST00000381908.3 ENST00000336164.4 ENST00000350274.5 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr6_-_116381918 | 0.03 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr1_+_197237352 | 0.02 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.5 | 3.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 2.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 3.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 3.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 3.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |