Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
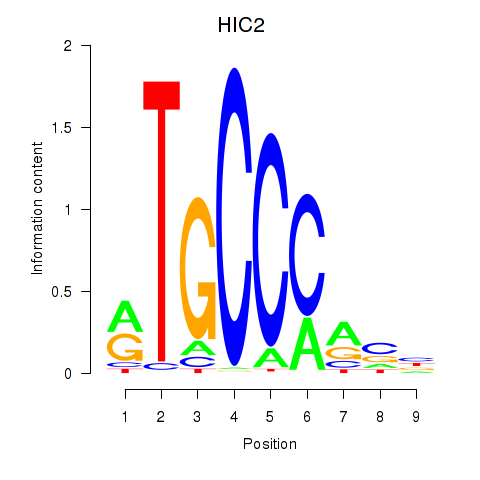
Results for HIC2
Z-value: 1.85
Transcription factors associated with HIC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIC2
|
ENSG00000169635.5 | HIC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIC2 | hg19_v2_chr22_+_21771656_21771693 | -0.35 | 1.8e-01 | Click! |
Activity profile of HIC2 motif
Sorted Z-values of HIC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_94856951 | 5.18 |
ENST00000553327.1 ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_94856987 | 4.76 |
ENST00000449399.3 ENST00000404814.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr17_-_39928106 | 4.47 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr16_+_56642489 | 3.97 |
ENST00000561491.1 |
MT2A |
metallothionein 2A |
| chr5_+_135385202 | 3.82 |
ENST00000514554.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr14_-_94857004 | 3.56 |
ENST00000557492.1 ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr10_-_91011548 | 3.44 |
ENST00000456827.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr2_-_224903995 | 3.42 |
ENST00000409304.1 ENST00000454956.1 ENST00000258405.4 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr2_-_161350305 | 3.23 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr6_-_132272504 | 3.00 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr9_+_124088860 | 2.92 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr10_+_124221036 | 2.87 |
ENST00000368984.3 |
HTRA1 |
HtrA serine peptidase 1 |
| chr2_+_173292280 | 2.79 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr16_-_18441131 | 2.74 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr18_+_21269404 | 2.68 |
ENST00000313654.9 |
LAMA3 |
laminin, alpha 3 |
| chr3_-_185542817 | 2.62 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr12_-_52845910 | 2.55 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr14_-_51411194 | 2.52 |
ENST00000544180.2 |
PYGL |
phosphorylase, glycogen, liver |
| chr12_-_52911718 | 2.50 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr2_+_173292301 | 2.50 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr22_+_33197683 | 2.49 |
ENST00000266085.6 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
| chr17_+_42427826 | 2.44 |
ENST00000586443.1 |
GRN |
granulin |
| chr10_+_124134201 | 2.41 |
ENST00000368990.3 ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr2_-_161349909 | 2.40 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr6_-_30712313 | 2.39 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr3_-_185542761 | 2.37 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr12_-_52967600 | 2.34 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr12_+_66217911 | 2.31 |
ENST00000403681.2 |
HMGA2 |
high mobility group AT-hook 2 |
| chr2_+_173292390 | 2.29 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr15_-_85201779 | 2.22 |
ENST00000360476.3 ENST00000394588.3 |
NMB |
neuromedin B |
| chr16_+_56659687 | 2.21 |
ENST00000568293.1 ENST00000330439.6 |
MT1E |
metallothionein 1E |
| chr19_-_51522955 | 2.21 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr3_-_99833333 | 2.20 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr2_+_235887329 | 2.19 |
ENST00000409212.1 ENST00000344528.4 ENST00000444916.1 |
SH3BP4 |
SH3-domain binding protein 4 |
| chr1_-_209825674 | 2.19 |
ENST00000367030.3 ENST00000356082.4 |
LAMB3 |
laminin, beta 3 |
| chr2_-_235405168 | 2.18 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr1_-_153363452 | 2.11 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr17_+_73717551 | 2.10 |
ENST00000450894.3 |
ITGB4 |
integrin, beta 4 |
| chr7_+_116165038 | 2.05 |
ENST00000393470.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr7_+_134212312 | 2.03 |
ENST00000359579.4 |
AKR1B10 |
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr18_+_21269556 | 2.02 |
ENST00000399516.3 |
LAMA3 |
laminin, alpha 3 |
| chr16_+_56642041 | 2.01 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr12_+_6309517 | 2.00 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr11_+_116700600 | 1.99 |
ENST00000227667.3 |
APOC3 |
apolipoprotein C-III |
| chr3_-_32022733 | 1.99 |
ENST00000438237.2 ENST00000396556.2 |
OSBPL10 |
oxysterol binding protein-like 10 |
| chr6_-_138428613 | 1.98 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chrX_+_114795489 | 1.98 |
ENST00000355899.3 ENST00000537301.1 ENST00000289290.3 |
PLS3 |
plastin 3 |
| chr11_-_2162468 | 1.95 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr7_+_79764104 | 1.95 |
ENST00000351004.3 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr16_-_11681316 | 1.95 |
ENST00000571688.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr11_+_116700614 | 1.94 |
ENST00000375345.1 |
APOC3 |
apolipoprotein C-III |
| chr6_+_31865552 | 1.94 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr8_-_125740730 | 1.90 |
ENST00000354184.4 |
MTSS1 |
metastasis suppressor 1 |
| chr9_+_124030338 | 1.88 |
ENST00000449773.1 ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN |
gelsolin |
| chr8_-_11710979 | 1.85 |
ENST00000415599.2 |
CTSB |
cathepsin B |
| chr10_-_125851961 | 1.85 |
ENST00000346248.5 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr1_-_153538292 | 1.84 |
ENST00000497140.1 ENST00000368708.3 |
S100A2 |
S100 calcium binding protein A2 |
| chr17_-_64216748 | 1.83 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_+_159570722 | 1.83 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr2_-_20424844 | 1.79 |
ENST00000403076.1 ENST00000254351.4 |
SDC1 |
syndecan 1 |
| chr7_-_107643674 | 1.78 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr6_+_151646800 | 1.77 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chrX_+_43515467 | 1.75 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr19_-_39264072 | 1.74 |
ENST00000599035.1 ENST00000378626.4 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
| chr10_-_126694575 | 1.73 |
ENST00000334808.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr2_-_238499303 | 1.73 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr1_+_32042131 | 1.70 |
ENST00000271064.7 ENST00000537531.1 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr11_-_2170786 | 1.69 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr5_+_52285144 | 1.67 |
ENST00000296585.5 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr1_-_153538011 | 1.67 |
ENST00000368707.4 |
S100A2 |
S100 calcium binding protein A2 |
| chr3_-_79068594 | 1.65 |
ENST00000436010.2 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chrX_-_131623982 | 1.63 |
ENST00000370844.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr1_-_237167718 | 1.63 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr12_-_53012343 | 1.62 |
ENST00000305748.3 |
KRT73 |
keratin 73 |
| chr1_+_32042105 | 1.61 |
ENST00000457433.2 ENST00000441210.2 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr13_-_45150392 | 1.61 |
ENST00000501704.2 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr10_-_17659357 | 1.59 |
ENST00000326961.6 ENST00000361271.3 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr3_+_105085734 | 1.56 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr18_+_21452964 | 1.55 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr4_+_4388805 | 1.55 |
ENST00000504171.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr3_-_149688502 | 1.55 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr2_-_70781087 | 1.54 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr7_+_116165754 | 1.54 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chrX_-_38080077 | 1.54 |
ENST00000378533.3 ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX |
sushi-repeat containing protein, X-linked |
| chr3_+_105086056 | 1.53 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr14_-_105420241 | 1.53 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr17_+_73717407 | 1.51 |
ENST00000579662.1 |
ITGB4 |
integrin, beta 4 |
| chr1_-_27240455 | 1.50 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr1_-_169555779 | 1.50 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr11_-_2162162 | 1.50 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr1_+_205473720 | 1.49 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr2_-_1748214 | 1.48 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr4_-_157892498 | 1.47 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr19_+_41725088 | 1.46 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr10_-_95360983 | 1.45 |
ENST00000371464.3 |
RBP4 |
retinol binding protein 4, plasma |
| chr3_-_149375783 | 1.43 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr1_+_19970797 | 1.43 |
ENST00000548815.1 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
| chr10_+_5566916 | 1.40 |
ENST00000315238.1 |
CALML3 |
calmodulin-like 3 |
| chr5_-_139726181 | 1.39 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr9_-_99381660 | 1.39 |
ENST00000375240.3 ENST00000463569.1 |
CDC14B |
cell division cycle 14B |
| chr3_-_50340996 | 1.38 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr10_-_33625154 | 1.37 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr11_+_10326612 | 1.34 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr17_+_42429493 | 1.34 |
ENST00000586242.1 |
GRN |
granulin |
| chr20_-_10654639 | 1.32 |
ENST00000254958.5 |
JAG1 |
jagged 1 |
| chr4_+_124317940 | 1.29 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr1_+_20915409 | 1.28 |
ENST00000375071.3 |
CDA |
cytidine deaminase |
| chr12_-_53009116 | 1.27 |
ENST00000552855.1 |
KRT73 |
keratin 73 |
| chr6_-_41715128 | 1.27 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr9_-_107690420 | 1.27 |
ENST00000423487.2 ENST00000374733.1 ENST00000374736.3 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr8_-_27457494 | 1.27 |
ENST00000521770.1 |
CLU |
clusterin |
| chrX_-_131623874 | 1.26 |
ENST00000436215.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr14_+_105939276 | 1.25 |
ENST00000483017.3 |
CRIP2 |
cysteine-rich protein 2 |
| chr2_-_88427568 | 1.24 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr1_+_19970657 | 1.23 |
ENST00000375136.3 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
| chr14_-_75422280 | 1.23 |
ENST00000238607.6 ENST00000553716.1 |
PGF |
placental growth factor |
| chr16_-_30122717 | 1.22 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr16_-_11681023 | 1.21 |
ENST00000570904.1 ENST00000574701.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr17_+_27369918 | 1.21 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chrX_-_43741594 | 1.20 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr20_+_61448376 | 1.20 |
ENST00000343916.3 |
COL9A3 |
collagen, type IX, alpha 3 |
| chr4_+_88896819 | 1.19 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr9_-_123812542 | 1.18 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr7_-_134143841 | 1.18 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr8_-_125740514 | 1.14 |
ENST00000325064.5 ENST00000518547.1 |
MTSS1 |
metastasis suppressor 1 |
| chr11_+_706113 | 1.13 |
ENST00000318562.8 ENST00000533256.1 ENST00000534755.1 |
EPS8L2 |
EPS8-like 2 |
| chr4_-_186733363 | 1.10 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_+_11674213 | 1.09 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr9_-_95896550 | 1.09 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr1_+_17944832 | 1.08 |
ENST00000167825.4 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr16_+_8891670 | 1.07 |
ENST00000268261.4 ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2 |
phosphomannomutase 2 |
| chr13_+_27131798 | 1.07 |
ENST00000361042.4 |
WASF3 |
WAS protein family, member 3 |
| chr7_-_27135591 | 1.06 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr12_-_15942503 | 1.06 |
ENST00000281172.5 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr2_-_56150184 | 1.05 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr2_+_187454749 | 1.04 |
ENST00000261023.3 ENST00000374907.3 |
ITGAV |
integrin, alpha V |
| chr7_-_27169801 | 1.04 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr19_-_51487282 | 1.03 |
ENST00000595820.1 ENST00000597707.1 ENST00000336317.4 |
KLK7 |
kallikrein-related peptidase 7 |
| chr17_+_48503603 | 1.03 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr9_-_99382065 | 1.03 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr17_-_48943706 | 1.02 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr7_-_27183263 | 1.02 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr9_+_19049372 | 1.02 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr12_-_52685312 | 1.01 |
ENST00000327741.5 |
KRT81 |
keratin 81 |
| chr1_+_17944806 | 1.00 |
ENST00000375408.3 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr9_-_112260531 | 0.99 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr10_-_99447024 | 0.99 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr12_+_6493199 | 0.99 |
ENST00000228918.4 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chrX_-_106362013 | 0.99 |
ENST00000372487.1 ENST00000372479.3 ENST00000203616.8 |
RBM41 |
RNA binding motif protein 41 |
| chr18_+_29171689 | 0.98 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr2_+_69001913 | 0.98 |
ENST00000409030.3 ENST00000409220.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_37571845 | 0.98 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chrX_-_153141302 | 0.97 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chr19_+_45449228 | 0.97 |
ENST00000252490.4 |
APOC2 |
apolipoprotein C-II |
| chr20_+_19867150 | 0.97 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr3_+_52828805 | 0.97 |
ENST00000416872.2 ENST00000449956.2 |
ITIH3 |
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr12_-_15942309 | 0.96 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr9_-_34637718 | 0.96 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr19_+_45449301 | 0.96 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr8_-_27472198 | 0.95 |
ENST00000519472.1 ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU |
clusterin |
| chr2_+_37571717 | 0.95 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr16_-_55867146 | 0.94 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr10_-_116286656 | 0.94 |
ENST00000428430.1 ENST00000369266.3 ENST00000392952.3 |
ABLIM1 |
actin binding LIM protein 1 |
| chr3_+_190231840 | 0.94 |
ENST00000072516.3 ENST00000443369.2 ENST00000412504.2 ENST00000439062.1 ENST00000447382.1 ENST00000422625.1 ENST00000422485.1 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chr11_-_66103867 | 0.94 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr4_+_169552748 | 0.94 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_+_100211412 | 0.94 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chrX_+_51927919 | 0.93 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr6_+_43739697 | 0.93 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr11_-_2158507 | 0.92 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chrX_+_95939638 | 0.92 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chrY_+_2709527 | 0.92 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr17_-_7017559 | 0.92 |
ENST00000446679.2 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr9_+_33750515 | 0.92 |
ENST00000361005.5 |
PRSS3 |
protease, serine, 3 |
| chr7_+_129932974 | 0.91 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr19_-_10697895 | 0.91 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr5_-_125930929 | 0.91 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr12_-_28125638 | 0.91 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr17_+_48503519 | 0.91 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr10_-_17659234 | 0.90 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr19_+_751122 | 0.90 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr16_-_87903079 | 0.90 |
ENST00000261622.4 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr2_-_106013400 | 0.89 |
ENST00000409807.1 |
FHL2 |
four and a half LIM domains 2 |
| chr12_-_85306594 | 0.88 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr21_+_46825032 | 0.88 |
ENST00000400337.2 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr2_-_235405679 | 0.88 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr11_+_47236489 | 0.88 |
ENST00000256996.4 ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2 |
damage-specific DNA binding protein 2, 48kDa |
| chr12_-_52828147 | 0.87 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr14_-_54421190 | 0.87 |
ENST00000417573.1 |
BMP4 |
bone morphogenetic protein 4 |
| chrX_-_153640420 | 0.87 |
ENST00000451865.1 ENST00000432135.1 ENST00000369809.1 ENST00000393638.1 ENST00000424626.1 ENST00000309585.5 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr10_+_102106829 | 0.87 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chrX_+_37208521 | 0.87 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr3_-_52479043 | 0.87 |
ENST00000231721.2 ENST00000475739.1 |
SEMA3G |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr16_-_66785497 | 0.86 |
ENST00000440564.2 ENST00000379482.2 ENST00000443351.2 ENST00000566150.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr17_+_73717516 | 0.86 |
ENST00000200181.3 ENST00000339591.3 |
ITGB4 |
integrin, beta 4 |
| chr2_+_70485220 | 0.85 |
ENST00000433351.2 ENST00000264441.5 |
PCYOX1 |
prenylcysteine oxidase 1 |
| chr17_-_7297519 | 0.85 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr22_-_37915247 | 0.85 |
ENST00000251973.5 |
CARD10 |
caspase recruitment domain family, member 10 |
| chrX_+_51636629 | 0.85 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr13_+_113633620 | 0.84 |
ENST00000421756.1 ENST00000375601.3 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.2 | 3.6 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.2 | 5.9 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.1 | 5.6 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.0 | 3.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.9 | 1.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.9 | 4.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 2.7 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.9 | 12.0 | GO:0035878 | nail development(GO:0035878) |
| 0.8 | 2.3 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.7 | 3.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.7 | 2.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.7 | 2.0 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.7 | 5.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 1.9 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.6 | 2.5 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.6 | 1.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.6 | 12.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 2.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.6 | 2.9 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.6 | 1.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.6 | 1.7 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 | 1.6 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.5 | 3.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.5 | 2.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 2.9 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 1.4 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.4 | 1.8 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.4 | 1.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.4 | 1.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.4 | 1.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.4 | 0.4 | GO:0030497 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation(GO:0030497) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.4 | 1.2 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 1.2 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.4 | 1.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.4 | 1.9 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.4 | 2.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 1.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.3 | 2.0 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 1.0 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.3 | 0.9 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.3 | 1.8 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.3 | 0.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 1.5 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.3 | 0.9 | GO:0061149 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 1.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 0.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.3 | 0.9 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 0.8 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.3 | 1.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 | 0.8 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.3 | 0.8 | GO:0098904 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) regulation of AV node cell action potential(GO:0098904) regulation of bundle of His cell action potential(GO:0098905) |
| 0.2 | 1.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.2 | 1.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.2 | 1.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 0.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 1.4 | GO:2000669 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.2 | 1.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.2 | 3.4 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.2 | 0.4 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.2 | 0.6 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 1.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.2 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 0.6 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.6 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 0.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.2 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.2 | 1.8 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 2.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 0.4 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.2 | 0.8 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 1.0 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 | 0.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 1.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.9 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 1.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) |
| 0.2 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 0.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 12.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.5 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.2 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 | 1.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 2.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.6 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.6 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 1.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 0.6 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 0.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.5 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 0.5 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 1.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.4 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.6 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 2.0 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 1.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.1 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.1 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.6 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.1 | 1.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 1.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.5 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 0.9 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.4 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.5 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 2.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.8 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.4 | GO:0014032 | neural crest cell development(GO:0014032) neural crest cell differentiation(GO:0014033) |
| 0.1 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 2.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.8 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 1.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.7 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 1.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.7 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.7 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 1.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.3 | GO:0070101 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.6 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.1 | 0.3 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 1.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.7 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 2.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 0.6 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 1.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.0 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.1 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.1 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) terpenoid catabolic process(GO:0016115) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.7 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.6 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 1.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 3.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.9 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.5 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 2.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.4 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 7.7 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.9 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.6 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 1.1 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.6 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 0.2 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 0.2 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 1.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.2 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 2.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 1.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.1 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.3 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.1 | 0.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:0072092 | olfactory bulb mitral cell layer development(GO:0061034) ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.1 | GO:2000418 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.1 | 1.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 1.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.7 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.1 | GO:0015793 | polyol transport(GO:0015791) glycerol transport(GO:0015793) |
| 0.1 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.1 | GO:0000726 | non-recombinational repair(GO:0000726) double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 1.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.1 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 3.6 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.3 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.5 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0045110 | neurofilament bundle assembly(GO:0033693) intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0042373 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.4 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0060688 | regulation of morphogenesis of a branching structure(GO:0060688) |
| 0.0 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.8 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0002903 | regulation of B cell apoptotic process(GO:0002902) negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.0 | 2.8 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.0 | 0.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0043500 | muscle adaptation(GO:0043500) |
| 0.0 | 0.2 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.3 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 1.6 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 1.0 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.0 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:1901070 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.5 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 1.0 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0097186 | amelogenesis(GO:0097186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 1.1 | 8.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.7 | 5.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.6 | 1.8 | GO:0043259 | laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.6 | 11.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.6 | 3.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 3.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 5.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 2.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 1.7 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.3 | 2.6 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 1.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 12.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 5.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 1.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 2.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.9 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 19.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.6 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 2.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.7 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 1.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 2.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 3.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.3 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 4.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 6.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 3.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 3.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 9.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 2.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 6.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 3.3 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.0 | 0.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.0 | 5.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 13.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.0 | 3.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.8 | 2.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.8 | 3.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.7 | 2.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.7 | 2.0 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.6 | 2.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 2.0 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.5 | 4.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.5 | 5.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 1.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 5.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.4 | 1.8 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 2.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.4 | 2.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 0.4 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.4 | 2.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 2.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 0.7 | GO:0050072 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) m7G(5')pppN diphosphatase activity(GO:0050072) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.4 | 2.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.4 | 2.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.3 | 0.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.3 | 1.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 2.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 3.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 1.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 0.8 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 6.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 0.6 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 1.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.8 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 1.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 20.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 0.5 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 0.7 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 0.5 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 7.4 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.2 | 1.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 0.7 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.2 | 0.7 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 1.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.5 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.2 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 5.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 4.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.1 | 0.9 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 1.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 2.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 4.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 1.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 3.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 5.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.4 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.3 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.0 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 12.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 22.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 11.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 13.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 13.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 16.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.4 | 8.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 6.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 7.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 19.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 2.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 5.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 12.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.3 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.1 | 2.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 4.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 4.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 3.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 3.8 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |