Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
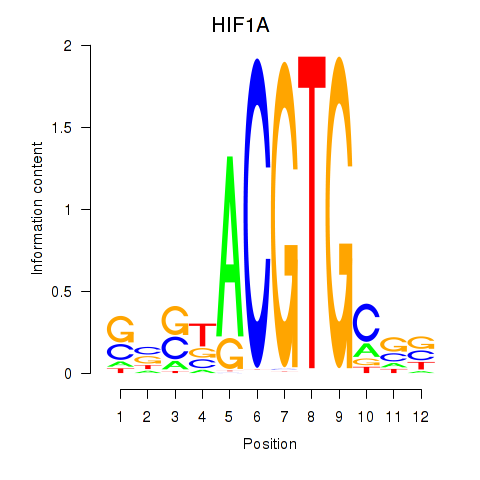
Results for HIF1A
Z-value: 1.13
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | HIF1A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg19_v2_chr14_+_62162258_62162269 | -0.19 | 4.8e-01 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_26331541 | 2.22 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr7_+_128095945 | 2.12 |
ENST00000257696.4 |
HILPDA |
hypoxia inducible lipid droplet-associated |
| chr14_+_64970662 | 1.84 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr12_+_7023491 | 1.83 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr15_+_89181974 | 1.77 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr14_-_64970494 | 1.74 |
ENST00000608382.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr15_+_89182178 | 1.73 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr12_+_7023735 | 1.72 |
ENST00000538763.1 ENST00000544774.1 ENST00000545045.2 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr15_+_89182156 | 1.67 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr2_-_216946500 | 1.63 |
ENST00000265322.7 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_+_65613852 | 1.63 |
ENST00000327299.7 |
AK4 |
adenylate kinase 4 |
| chrX_+_24483338 | 1.60 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr15_-_60884706 | 1.59 |
ENST00000449337.2 |
RORA |
RAR-related orphan receptor A |
| chr1_-_92351769 | 1.44 |
ENST00000212355.4 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr6_+_89791507 | 1.43 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr11_+_7534999 | 1.37 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_88179302 | 1.29 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr12_+_6977258 | 1.24 |
ENST00000488464.2 ENST00000535434.1 ENST00000493987.1 |
TPI1 |
triosephosphate isomerase 1 |
| chr19_+_41903709 | 1.18 |
ENST00000542943.1 ENST00000457836.2 |
BCKDHA |
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr5_-_88178964 | 1.14 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr3_-_195808952 | 1.10 |
ENST00000540528.1 ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC |
transferrin receptor |
| chr2_+_173420697 | 1.04 |
ENST00000282077.3 ENST00000392571.2 ENST00000410055.1 |
PDK1 |
pyruvate dehydrogenase kinase, isozyme 1 |
| chr5_-_88179017 | 1.03 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr9_+_4490394 | 1.01 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr5_+_49962772 | 0.97 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr21_+_45719921 | 0.96 |
ENST00000349048.4 |
PFKL |
phosphofructokinase, liver |
| chr3_-_195808980 | 0.96 |
ENST00000360110.4 |
TFRC |
transferrin receptor |
| chr2_+_118846008 | 0.93 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr1_-_151319283 | 0.92 |
ENST00000392746.3 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr2_-_220083076 | 0.91 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr17_+_5185552 | 0.90 |
ENST00000262477.6 ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
| chr9_+_117373486 | 0.87 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr15_+_41056255 | 0.85 |
ENST00000561160.1 ENST00000559445.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr19_-_5719860 | 0.85 |
ENST00000590729.1 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr6_-_153304148 | 0.84 |
ENST00000229758.3 |
FBXO5 |
F-box protein 5 |
| chr2_-_214016314 | 0.82 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr15_+_41056218 | 0.81 |
ENST00000260447.4 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr19_-_41903161 | 0.80 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr1_-_8939265 | 0.79 |
ENST00000489867.1 |
ENO1 |
enolase 1, (alpha) |
| chr1_-_32801825 | 0.79 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr19_-_5720123 | 0.78 |
ENST00000587365.1 ENST00000585374.1 ENST00000593119.1 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr17_+_65373531 | 0.75 |
ENST00000580974.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr20_+_30946106 | 0.71 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr8_+_98656336 | 0.71 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr19_-_5720248 | 0.69 |
ENST00000360614.3 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr8_+_26240414 | 0.68 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_-_103424916 | 0.67 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr5_+_126112794 | 0.67 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr6_+_71377473 | 0.66 |
ENST00000370452.3 ENST00000316999.5 ENST00000370455.3 |
SMAP1 |
small ArfGAP 1 |
| chr17_-_35969409 | 0.66 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr17_-_67323305 | 0.66 |
ENST00000392677.2 ENST00000593153.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr8_-_103425047 | 0.65 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chrX_+_77359726 | 0.64 |
ENST00000442431.1 |
PGK1 |
phosphoglycerate kinase 1 |
| chr17_-_67323232 | 0.64 |
ENST00000592568.1 ENST00000392676.3 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr3_+_133292851 | 0.64 |
ENST00000503932.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr6_-_153304697 | 0.63 |
ENST00000367241.3 |
FBXO5 |
F-box protein 5 |
| chr6_-_112194484 | 0.63 |
ENST00000518295.1 ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr8_+_98656693 | 0.61 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr17_+_80416482 | 0.61 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chr5_+_138609441 | 0.60 |
ENST00000509990.1 ENST00000506147.1 ENST00000512107.1 |
MATR3 |
matrin 3 |
| chr8_-_103424986 | 0.60 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chrX_+_77359671 | 0.59 |
ENST00000373316.4 |
PGK1 |
phosphoglycerate kinase 1 |
| chr17_+_80416050 | 0.59 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr15_+_74908147 | 0.58 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr2_+_88991162 | 0.58 |
ENST00000283646.4 |
RPIA |
ribose 5-phosphate isomerase A |
| chr4_+_17578815 | 0.57 |
ENST00000226299.4 |
LAP3 |
leucine aminopeptidase 3 |
| chr14_-_92572894 | 0.57 |
ENST00000532032.1 ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3 |
ataxin 3 |
| chr7_+_116593433 | 0.57 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr12_+_72148614 | 0.56 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr19_-_49137790 | 0.55 |
ENST00000599385.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr13_+_103451399 | 0.55 |
ENST00000257336.1 ENST00000448849.2 |
BIVM |
basic, immunoglobulin-like variable motif containing |
| chr2_+_204193149 | 0.53 |
ENST00000422511.2 |
ABI2 |
abl-interactor 2 |
| chr7_+_94537542 | 0.51 |
ENST00000433881.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr6_+_87865262 | 0.51 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr3_-_81811312 | 0.51 |
ENST00000429644.2 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr3_+_133292759 | 0.50 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chr17_-_74449252 | 0.50 |
ENST00000319380.7 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
| chr8_+_28351707 | 0.49 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr2_-_220083692 | 0.48 |
ENST00000265316.3 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr13_-_52027134 | 0.48 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr12_+_110906169 | 0.47 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr14_-_53162361 | 0.46 |
ENST00000395686.3 |
ERO1L |
ERO1-like (S. cerevisiae) |
| chr5_+_49962495 | 0.46 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_112162381 | 0.45 |
ENST00000433097.1 ENST00000369709.3 ENST00000436150.2 |
RAP1A |
RAP1A, member of RAS oncogene family |
| chr9_+_100745615 | 0.45 |
ENST00000339399.4 |
ANP32B |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr2_-_220083671 | 0.44 |
ENST00000439002.2 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr15_-_34875771 | 0.44 |
ENST00000267731.7 |
GOLGA8B |
golgin A8 family, member B |
| chr12_+_6643676 | 0.41 |
ENST00000396856.1 ENST00000396861.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_204192942 | 0.41 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chrX_+_69509927 | 0.41 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr4_+_76649797 | 0.41 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr6_+_108487245 | 0.40 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr17_+_72428218 | 0.40 |
ENST00000392628.2 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_204193129 | 0.40 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr6_+_64282447 | 0.39 |
ENST00000370650.2 ENST00000578299.1 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr1_-_8938736 | 0.38 |
ENST00000234590.4 |
ENO1 |
enolase 1, (alpha) |
| chr1_-_51425772 | 0.38 |
ENST00000371778.4 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr8_-_95908902 | 0.37 |
ENST00000520509.1 |
CCNE2 |
cyclin E2 |
| chr19_-_49137762 | 0.37 |
ENST00000593500.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr11_-_57283159 | 0.36 |
ENST00000533263.1 ENST00000278426.3 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr7_+_96747030 | 0.36 |
ENST00000360382.4 |
ACN9 |
ACN9 homolog (S. cerevisiae) |
| chr5_+_70883117 | 0.36 |
ENST00000340941.6 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr12_-_121734489 | 0.35 |
ENST00000412367.2 ENST00000402834.4 ENST00000404169.3 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr11_-_118901559 | 0.35 |
ENST00000330775.7 ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr3_+_184080790 | 0.34 |
ENST00000430783.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_45805667 | 0.34 |
ENST00000488731.2 ENST00000435155.1 |
MUTYH |
mutY homolog |
| chr2_+_216176761 | 0.34 |
ENST00000540518.1 ENST00000435675.1 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr7_-_22396533 | 0.34 |
ENST00000344041.6 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr22_+_39853258 | 0.34 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr1_-_6259641 | 0.34 |
ENST00000234875.4 |
RPL22 |
ribosomal protein L22 |
| chr5_+_70883178 | 0.34 |
ENST00000323375.8 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr13_+_25875785 | 0.34 |
ENST00000381747.3 |
NUPL1 |
nucleoporin like 1 |
| chr5_+_61602055 | 0.34 |
ENST00000381103.2 |
KIF2A |
kinesin heavy chain member 2A |
| chr16_+_16043406 | 0.34 |
ENST00000399410.3 ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr1_-_16678914 | 0.34 |
ENST00000375592.3 |
FBXO42 |
F-box protein 42 |
| chr1_+_110162448 | 0.33 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr5_+_176853702 | 0.33 |
ENST00000507633.1 ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6 |
G protein-coupled receptor kinase 6 |
| chr1_+_231376941 | 0.32 |
ENST00000436239.1 ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT |
glyceronephosphate O-acyltransferase |
| chr1_-_6269448 | 0.32 |
ENST00000465335.1 |
RPL22 |
ribosomal protein L22 |
| chr12_+_6644443 | 0.32 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_+_31477250 | 0.32 |
ENST00000313737.4 |
AC024940.1 |
AC024940.1 |
| chr10_-_96122682 | 0.32 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr7_+_94536898 | 0.31 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_216176540 | 0.31 |
ENST00000236959.9 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr14_+_77787227 | 0.31 |
ENST00000216465.5 ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr1_-_45805607 | 0.31 |
ENST00000372104.1 ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH |
mutY homolog |
| chr18_-_45457478 | 0.30 |
ENST00000402690.2 ENST00000356825.4 |
SMAD2 |
SMAD family member 2 |
| chr3_-_49377499 | 0.30 |
ENST00000265560.4 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr13_-_24463530 | 0.30 |
ENST00000382172.3 |
MIPEP |
mitochondrial intermediate peptidase |
| chr15_-_74726283 | 0.30 |
ENST00000543145.2 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr8_+_17354587 | 0.29 |
ENST00000494857.1 ENST00000522656.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr7_-_139876812 | 0.28 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr19_+_38397839 | 0.28 |
ENST00000222345.6 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
| chr21_-_38639601 | 0.28 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr3_-_183543301 | 0.28 |
ENST00000318631.3 ENST00000431348.1 |
MAP6D1 |
MAP6 domain containing 1 |
| chr12_-_49318715 | 0.28 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr6_-_151773232 | 0.28 |
ENST00000444024.1 ENST00000367303.4 |
RMND1 |
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr6_+_35265586 | 0.27 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr8_+_17354617 | 0.27 |
ENST00000470360.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr6_+_64281906 | 0.27 |
ENST00000370651.3 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr6_-_43021612 | 0.27 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
| chr3_-_49377446 | 0.27 |
ENST00000351842.4 ENST00000416417.1 ENST00000415188.1 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr13_-_44361025 | 0.27 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chr22_-_36903101 | 0.27 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr11_-_73471655 | 0.26 |
ENST00000400470.2 |
RAB6A |
RAB6A, member RAS oncogene family |
| chr1_+_228353495 | 0.26 |
ENST00000366711.3 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr19_+_50180409 | 0.26 |
ENST00000391851.4 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr22_-_31741757 | 0.26 |
ENST00000215919.3 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_-_6269304 | 0.25 |
ENST00000471204.1 |
RPL22 |
ribosomal protein L22 |
| chr1_-_115053781 | 0.24 |
ENST00000358465.2 ENST00000369543.2 |
TRIM33 |
tripartite motif containing 33 |
| chr7_-_92465868 | 0.24 |
ENST00000424848.2 |
CDK6 |
cyclin-dependent kinase 6 |
| chr6_+_127588020 | 0.24 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr9_+_133884469 | 0.24 |
ENST00000361069.4 |
LAMC3 |
laminin, gamma 3 |
| chr11_-_35440579 | 0.24 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_+_64564469 | 0.24 |
ENST00000373783.1 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
| chr22_-_36903069 | 0.24 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr19_+_50180507 | 0.23 |
ENST00000454376.2 ENST00000524771.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr14_-_64010046 | 0.23 |
ENST00000337537.3 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr11_-_76155700 | 0.23 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr1_+_193091080 | 0.23 |
ENST00000367435.3 |
CDC73 |
cell division cycle 73 |
| chr8_-_117768023 | 0.23 |
ENST00000518949.1 ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
| chr22_+_24236191 | 0.22 |
ENST00000215754.7 |
MIF |
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr11_-_116968987 | 0.22 |
ENST00000434315.2 ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3 |
SIK family kinase 3 |
| chr7_+_128095900 | 0.22 |
ENST00000435296.2 |
HILPDA |
hypoxia inducible lipid droplet-associated |
| chr9_-_139922631 | 0.21 |
ENST00000341511.6 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_73498898 | 0.21 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chr17_+_25621102 | 0.20 |
ENST00000581440.1 ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1 |
WD repeat and SOCS box containing 1 |
| chr9_+_133569108 | 0.20 |
ENST00000372358.5 ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2 |
exosome component 2 |
| chr11_+_14665263 | 0.20 |
ENST00000282096.4 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
| chr20_+_48552908 | 0.19 |
ENST00000244061.2 |
RNF114 |
ring finger protein 114 |
| chr16_+_69166418 | 0.19 |
ENST00000314423.7 ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A |
cirrhosis, autosomal recessive 1A (cirhin) |
| chr2_+_32288725 | 0.19 |
ENST00000315285.3 |
SPAST |
spastin |
| chr1_-_1709845 | 0.19 |
ENST00000341426.5 ENST00000344463.4 |
NADK |
NAD kinase |
| chr11_-_35440796 | 0.19 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_-_39529180 | 0.18 |
ENST00000515021.1 ENST00000510490.1 ENST00000316423.6 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr2_-_40006289 | 0.18 |
ENST00000260619.6 ENST00000454352.2 |
THUMPD2 |
THUMP domain containing 2 |
| chr2_+_32288657 | 0.18 |
ENST00000345662.1 |
SPAST |
spastin |
| chr12_-_113909877 | 0.17 |
ENST00000261731.3 |
LHX5 |
LIM homeobox 5 |
| chrX_-_47509887 | 0.17 |
ENST00000247161.3 ENST00000592066.1 ENST00000376983.3 |
ELK1 |
ELK1, member of ETS oncogene family |
| chr11_-_18034701 | 0.17 |
ENST00000265965.5 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr8_-_30515693 | 0.17 |
ENST00000355904.4 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr8_-_101734907 | 0.17 |
ENST00000318607.5 ENST00000521865.1 ENST00000520804.1 ENST00000522720.1 ENST00000521067.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr14_-_23770683 | 0.16 |
ENST00000561437.1 ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E |
protein phosphatase 1, regulatory subunit 3E |
| chr2_-_28113965 | 0.16 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chr17_+_76311791 | 0.16 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr1_-_6259613 | 0.16 |
ENST00000465387.1 |
RPL22 |
ribosomal protein L22 |
| chr11_-_76155618 | 0.16 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr14_-_54908043 | 0.16 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr8_+_97274119 | 0.16 |
ENST00000455950.2 |
PTDSS1 |
phosphatidylserine synthase 1 |
| chr4_+_78079450 | 0.16 |
ENST00000395640.1 ENST00000512918.1 |
CCNG2 |
cyclin G2 |
| chr6_-_109702885 | 0.15 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr21_+_45138941 | 0.15 |
ENST00000398081.1 ENST00000468090.1 ENST00000291565.4 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr11_-_100999775 | 0.14 |
ENST00000263463.5 |
PGR |
progesterone receptor |
| chr4_-_39529049 | 0.14 |
ENST00000501493.2 ENST00000509391.1 ENST00000507089.1 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr22_-_38577731 | 0.14 |
ENST00000335539.3 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr15_+_52311398 | 0.14 |
ENST00000261845.5 |
MAPK6 |
mitogen-activated protein kinase 6 |
| chr16_+_53164833 | 0.14 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr3_+_52719936 | 0.14 |
ENST00000418458.1 ENST00000394799.2 |
GNL3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr12_-_123849374 | 0.14 |
ENST00000602398.1 ENST00000602750.1 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
| chr1_-_151319710 | 0.14 |
ENST00000290524.4 ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 2.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 2.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 5.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 2.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 4.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 3.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.8 | 2.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.6 | 2.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 1.7 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.5 | 1.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.5 | 1.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.5 | 1.8 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.4 | 1.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 2.6 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 1.0 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 1.9 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 1.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.3 | 0.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 0.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.7 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.8 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 0.6 | GO:1903401 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transmembrane transport(GO:1903401) L-lysine import into cell(GO:1903410) |
| 0.2 | 1.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 1.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.5 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.2 | 1.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 0.7 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.2 | 0.5 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.6 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 5.7 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.8 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 2.1 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.5 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.7 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 2.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.3 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 1.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.2 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.3 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0060748 | paracrine signaling(GO:0038001) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.0 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 2.3 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.7 | 2.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.7 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 1.8 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.6 | 1.7 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.5 | 1.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 1.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 1.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.4 | 1.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.2 | 1.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 3.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 0.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 0.7 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 1.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 1.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.5 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.6 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0016413 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 6.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |