Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
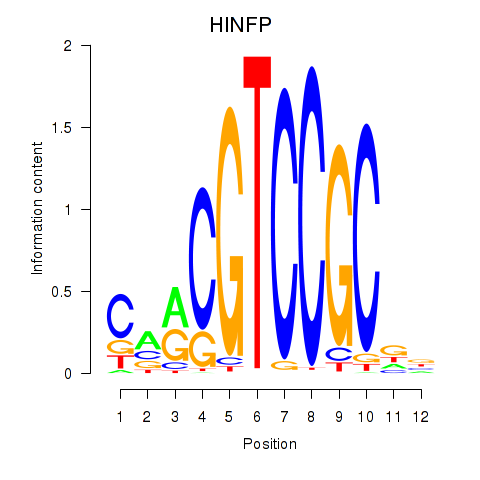
Results for HINFP
Z-value: 0.57
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | HINFP |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.24 | 3.8e-01 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_94023873 | 2.13 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr5_-_136834982 | 1.04 |
ENST00000510689.1 ENST00000394945.1 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr11_-_2162468 | 0.98 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_110790590 | 0.98 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr11_-_2162162 | 0.96 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr10_-_62761188 | 0.92 |
ENST00000357917.4 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr9_+_137533615 | 0.92 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr3_-_38691119 | 0.86 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr12_-_90102594 | 0.85 |
ENST00000428670.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_-_186733363 | 0.79 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr3_-_16555150 | 0.67 |
ENST00000334133.4 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr17_-_46115122 | 0.62 |
ENST00000006101.4 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
| chr6_-_139695757 | 0.57 |
ENST00000367651.2 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr16_+_6069586 | 0.48 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr13_-_77460525 | 0.47 |
ENST00000377474.2 ENST00000317765.2 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
| chr3_+_49449636 | 0.45 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr9_-_89562104 | 0.41 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr15_+_43803143 | 0.41 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr13_-_36705425 | 0.39 |
ENST00000255448.4 ENST00000360631.3 ENST00000379892.4 |
DCLK1 |
doublecortin-like kinase 1 |
| chr11_-_66336060 | 0.34 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr11_-_72353451 | 0.33 |
ENST00000376450.3 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr1_+_204797749 | 0.33 |
ENST00000367172.4 ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC |
neurofascin |
| chr5_+_72143988 | 0.29 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr1_-_94703118 | 0.29 |
ENST00000260526.6 ENST00000370217.3 |
ARHGAP29 |
Rho GTPase activating protein 29 |
| chr17_-_1419878 | 0.28 |
ENST00000449479.1 ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr8_-_27462822 | 0.27 |
ENST00000522098.1 |
CLU |
clusterin |
| chr4_+_123747834 | 0.27 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr3_-_49449521 | 0.26 |
ENST00000431929.1 ENST00000418115.1 |
RHOA |
ras homolog family member A |
| chr13_+_25875785 | 0.26 |
ENST00000381747.3 |
NUPL1 |
nucleoporin like 1 |
| chr13_-_41240717 | 0.26 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr17_-_1419914 | 0.25 |
ENST00000397335.3 ENST00000574561.1 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr13_+_111806055 | 0.25 |
ENST00000218789.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr18_-_44336998 | 0.24 |
ENST00000315087.7 |
ST8SIA5 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr3_-_129612394 | 0.24 |
ENST00000505616.1 ENST00000426664.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr17_+_80186908 | 0.23 |
ENST00000582743.1 ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr7_-_134143841 | 0.23 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr3_-_49449350 | 0.22 |
ENST00000454011.2 ENST00000445425.1 ENST00000422781.1 |
RHOA |
ras homolog family member A |
| chrX_-_119695279 | 0.22 |
ENST00000336592.6 |
CUL4B |
cullin 4B |
| chr17_-_1420182 | 0.22 |
ENST00000421807.2 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr8_+_42995548 | 0.22 |
ENST00000458501.2 ENST00000379644.4 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
| chr13_-_22178284 | 0.21 |
ENST00000468222.2 ENST00000382374.4 |
MICU2 |
mitochondrial calcium uptake 2 |
| chr3_+_139654018 | 0.20 |
ENST00000458420.3 |
CLSTN2 |
calsyntenin 2 |
| chr1_+_93913713 | 0.20 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chrX_-_38186811 | 0.20 |
ENST00000318842.7 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr17_+_46125707 | 0.19 |
ENST00000584137.1 ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr13_+_25875662 | 0.19 |
ENST00000381736.3 ENST00000463407.1 ENST00000381718.3 |
NUPL1 |
nucleoporin like 1 |
| chr3_-_49377446 | 0.19 |
ENST00000351842.4 ENST00000416417.1 ENST00000415188.1 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr12_+_102271129 | 0.19 |
ENST00000258534.8 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
| chr3_-_49377499 | 0.18 |
ENST00000265560.4 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr3_-_156272924 | 0.18 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr11_+_62104897 | 0.18 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr2_+_10442993 | 0.18 |
ENST00000423674.1 ENST00000307845.3 |
HPCAL1 |
hippocalcin-like 1 |
| chr17_-_1420006 | 0.17 |
ENST00000320345.6 ENST00000406424.4 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr6_+_42847348 | 0.17 |
ENST00000493763.1 ENST00000304734.5 |
RPL7L1 |
ribosomal protein L7-like 1 |
| chr10_-_118502070 | 0.16 |
ENST00000369209.3 |
HSPA12A |
heat shock 70kDa protein 12A |
| chr13_+_111806083 | 0.16 |
ENST00000375736.4 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_-_66453562 | 0.15 |
ENST00000262139.5 ENST00000546360.1 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
| chr11_-_6633799 | 0.15 |
ENST00000299424.4 |
TAF10 |
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr3_-_57583185 | 0.15 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr4_+_123747979 | 0.15 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr2_+_204193149 | 0.15 |
ENST00000422511.2 |
ABI2 |
abl-interactor 2 |
| chr12_+_21654714 | 0.15 |
ENST00000542038.1 ENST00000540141.1 ENST00000229314.5 |
GOLT1B |
golgi transport 1B |
| chr5_+_14143728 | 0.15 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chrX_+_40944871 | 0.15 |
ENST00000378308.2 ENST00000324545.8 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
| chrX_-_153602991 | 0.15 |
ENST00000369850.3 ENST00000422373.1 |
FLNA |
filamin A, alpha |
| chr8_+_6565854 | 0.14 |
ENST00000285518.6 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr17_-_42277203 | 0.14 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr11_-_6440283 | 0.14 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chrX_-_38186775 | 0.14 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr17_+_46126135 | 0.14 |
ENST00000361665.3 ENST00000585062.1 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr13_+_21714653 | 0.14 |
ENST00000382533.4 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr6_-_111804905 | 0.13 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr13_+_21714711 | 0.13 |
ENST00000607003.1 ENST00000492245.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr22_+_18593446 | 0.13 |
ENST00000316027.6 |
TUBA8 |
tubulin, alpha 8 |
| chr2_+_120770645 | 0.13 |
ENST00000443902.2 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_204193129 | 0.13 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr2_-_175547571 | 0.12 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr4_+_166128735 | 0.12 |
ENST00000226725.6 |
KLHL2 |
kelch-like family member 2 |
| chr2_+_120770581 | 0.12 |
ENST00000263713.5 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr11_-_6440624 | 0.12 |
ENST00000311051.3 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_+_42086525 | 0.12 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr11_+_125462690 | 0.12 |
ENST00000392708.4 ENST00000529196.1 ENST00000531491.1 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr8_+_26240414 | 0.11 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_+_64971292 | 0.11 |
ENST00000358738.3 ENST00000394712.2 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr11_-_62380199 | 0.10 |
ENST00000419857.1 ENST00000394773.2 |
EML3 |
echinoderm microtubule associated protein like 3 |
| chr4_-_149363662 | 0.10 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr3_-_57583052 | 0.09 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr20_+_57267669 | 0.09 |
ENST00000356091.6 |
NPEPL1 |
aminopeptidase-like 1 |
| chr19_-_14016877 | 0.09 |
ENST00000454313.1 ENST00000591586.1 ENST00000346736.2 |
C19orf57 |
chromosome 19 open reading frame 57 |
| chr16_+_81069433 | 0.08 |
ENST00000299575.4 |
ATMIN |
ATM interactor |
| chr10_-_135187193 | 0.08 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr15_-_43802769 | 0.08 |
ENST00000263801.3 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr3_-_57583130 | 0.07 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr4_-_183838372 | 0.07 |
ENST00000503820.1 ENST00000503988.1 |
DCTD |
dCMP deaminase |
| chr2_+_121010324 | 0.07 |
ENST00000272519.5 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
| chr14_-_103523745 | 0.06 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr2_-_232791038 | 0.06 |
ENST00000295440.2 ENST00000409852.1 |
NPPC |
natriuretic peptide C |
| chr13_+_21714913 | 0.06 |
ENST00000450573.1 ENST00000467636.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr4_-_183838596 | 0.06 |
ENST00000508994.1 ENST00000512766.1 |
DCTD |
dCMP deaminase |
| chr2_+_121010370 | 0.06 |
ENST00000420510.1 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
| chr3_-_49131473 | 0.06 |
ENST00000430979.1 ENST00000357496.2 ENST00000437939.1 |
QRICH1 |
glutamine-rich 1 |
| chr13_+_48877895 | 0.06 |
ENST00000267163.4 |
RB1 |
retinoblastoma 1 |
| chr20_+_30946106 | 0.06 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr9_-_122131696 | 0.06 |
ENST00000373964.2 ENST00000265922.3 |
BRINP1 |
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr2_+_121010413 | 0.06 |
ENST00000404963.3 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
| chr8_+_32406179 | 0.05 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr15_-_35047166 | 0.05 |
ENST00000290374.4 |
GJD2 |
gap junction protein, delta 2, 36kDa |
| chr10_+_94352956 | 0.05 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr5_+_142149932 | 0.05 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr2_-_33824382 | 0.05 |
ENST00000238823.8 |
FAM98A |
family with sequence similarity 98, member A |
| chr1_+_29063119 | 0.05 |
ENST00000474884.1 ENST00000542507.1 |
YTHDF2 |
YTH domain family, member 2 |
| chr12_+_113659234 | 0.04 |
ENST00000551096.1 ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1 |
two pore segment channel 1 |
| chr19_-_55919087 | 0.04 |
ENST00000587845.1 ENST00000589978.1 ENST00000264552.9 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
| chr19_-_14117074 | 0.04 |
ENST00000588885.1 ENST00000254325.4 |
RFX1 |
regulatory factor X, 1 (influences HLA class II expression) |
| chr5_+_61602055 | 0.04 |
ENST00000381103.2 |
KIF2A |
kinesin heavy chain member 2A |
| chr14_-_92572894 | 0.04 |
ENST00000532032.1 ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3 |
ataxin 3 |
| chr17_-_7518145 | 0.04 |
ENST00000250113.7 ENST00000571597.1 |
FXR2 |
fragile X mental retardation, autosomal homolog 2 |
| chr9_-_139094988 | 0.04 |
ENST00000371746.3 |
LHX3 |
LIM homeobox 3 |
| chr17_-_73178599 | 0.04 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr19_-_39881669 | 0.04 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr5_+_142149955 | 0.04 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr1_+_29063439 | 0.04 |
ENST00000541996.1 ENST00000496288.1 |
YTHDF2 |
YTH domain family, member 2 |
| chr5_+_113697983 | 0.04 |
ENST00000264773.3 |
KCNN2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr22_-_43010928 | 0.04 |
ENST00000348657.2 ENST00000252115.5 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_-_70557430 | 0.04 |
ENST00000393612.4 ENST00000564653.1 ENST00000323786.5 |
COG4 |
component of oligomeric golgi complex 4 |
| chr7_-_99006443 | 0.04 |
ENST00000350498.3 |
PDAP1 |
PDGFA associated protein 1 |
| chr5_-_132166579 | 0.04 |
ENST00000378679.3 |
SHROOM1 |
shroom family member 1 |
| chr6_-_35464817 | 0.03 |
ENST00000338863.7 |
TEAD3 |
TEA domain family member 3 |
| chr1_+_22351977 | 0.03 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr7_-_26240357 | 0.03 |
ENST00000354667.4 ENST00000356674.7 |
HNRNPA2B1 |
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr20_-_39946237 | 0.03 |
ENST00000441102.2 ENST00000559234.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr2_-_33824336 | 0.03 |
ENST00000431950.1 ENST00000403368.1 ENST00000441530.2 |
FAM98A |
family with sequence similarity 98, member A |
| chr9_+_100174344 | 0.03 |
ENST00000422139.2 |
TDRD7 |
tudor domain containing 7 |
| chr17_-_4544960 | 0.03 |
ENST00000293761.3 |
ALOX15 |
arachidonate 15-lipoxygenase |
| chr17_+_43971643 | 0.03 |
ENST00000344290.5 ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT |
microtubule-associated protein tau |
| chr6_+_42847649 | 0.03 |
ENST00000424341.2 ENST00000602561.1 |
RPL7L1 |
ribosomal protein L7-like 1 |
| chr1_+_29063271 | 0.03 |
ENST00000373812.3 |
YTHDF2 |
YTH domain family, member 2 |
| chr3_-_49761337 | 0.03 |
ENST00000535833.1 ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3 GMPPB |
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr9_+_77112244 | 0.03 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr9_-_139922726 | 0.02 |
ENST00000265662.5 ENST00000371605.3 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr10_+_99079008 | 0.02 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr7_+_101459263 | 0.02 |
ENST00000292538.4 ENST00000393824.3 ENST00000547394.2 ENST00000360264.3 ENST00000425244.2 |
CUX1 |
cut-like homeobox 1 |
| chr9_+_34458771 | 0.02 |
ENST00000437363.1 ENST00000242317.4 |
DNAI1 |
dynein, axonemal, intermediate chain 1 |
| chr6_-_35464727 | 0.02 |
ENST00000402886.3 |
TEAD3 |
TEA domain family member 3 |
| chr1_-_197115818 | 0.02 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr14_-_99947168 | 0.02 |
ENST00000331768.5 |
SETD3 |
SET domain containing 3 |
| chr11_-_73309228 | 0.02 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr5_+_176873446 | 0.02 |
ENST00000507881.1 |
PRR7 |
proline rich 7 (synaptic) |
| chr5_-_114598548 | 0.02 |
ENST00000379615.3 ENST00000419445.1 |
PGGT1B |
protein geranylgeranyltransferase type I, beta subunit |
| chr8_-_100905363 | 0.02 |
ENST00000524245.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr1_-_40157345 | 0.02 |
ENST00000372844.3 |
HPCAL4 |
hippocalcin like 4 |
| chrX_+_21392553 | 0.02 |
ENST00000279451.4 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_97304105 | 0.02 |
ENST00000599854.1 ENST00000441706.2 |
KANSL3 |
KAT8 regulatory NSL complex subunit 3 |
| chr6_-_144329531 | 0.02 |
ENST00000429150.1 ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr8_-_30670053 | 0.02 |
ENST00000518564.1 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr4_-_2758015 | 0.01 |
ENST00000510267.1 ENST00000503235.1 ENST00000315423.7 |
TNIP2 |
TNFAIP3 interacting protein 2 |
| chr19_-_39881777 | 0.01 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr4_-_183838482 | 0.01 |
ENST00000357067.3 ENST00000510370.1 |
DCTD |
dCMP deaminase |
| chr17_+_43972010 | 0.01 |
ENST00000334239.8 ENST00000446361.3 |
MAPT |
microtubule-associated protein tau |
| chr16_+_56899114 | 0.01 |
ENST00000566786.1 ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3 |
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr2_+_64068074 | 0.01 |
ENST00000394417.2 ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr19_-_45004556 | 0.01 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr3_-_48936272 | 0.01 |
ENST00000544097.1 ENST00000430379.1 ENST00000319017.4 |
SLC25A20 |
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr12_+_51985001 | 0.01 |
ENST00000354534.6 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
| chr4_-_156298028 | 0.01 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr1_+_6845384 | 0.01 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr4_-_183838747 | 0.01 |
ENST00000438320.2 |
DCTD |
dCMP deaminase |
| chr11_+_28131821 | 0.00 |
ENST00000379199.2 ENST00000303459.6 |
METTL15 |
methyltransferase like 15 |
| chr19_-_10530784 | 0.00 |
ENST00000593124.1 |
CDC37 |
cell division cycle 37 |
| chr2_+_157292859 | 0.00 |
ENST00000438166.2 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr2_+_42396574 | 0.00 |
ENST00000401738.3 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr2_-_170219079 | 0.00 |
ENST00000263816.3 |
LRP2 |
low density lipoprotein receptor-related protein 2 |
| chr12_+_57482665 | 0.00 |
ENST00000300131.3 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr11_-_62446527 | 0.00 |
ENST00000294119.2 ENST00000529640.1 ENST00000534176.1 ENST00000301935.5 |
UBXN1 |
UBX domain protein 1 |
| chr12_+_70637494 | 0.00 |
ENST00000548159.1 ENST00000549750.1 ENST00000551043.1 |
CNOT2 |
CCR4-NOT transcription complex, subunit 2 |
| chr6_-_33267101 | 0.00 |
ENST00000497454.1 |
RGL2 |
ral guanine nucleotide dissociation stimulator-like 2 |
| chr7_+_128095900 | 0.00 |
ENST00000435296.2 |
HILPDA |
hypoxia inducible lipid droplet-associated |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 0.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.2 | 1.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.9 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 2.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.6 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.5 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.5 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.7 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.3 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 1.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |