Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
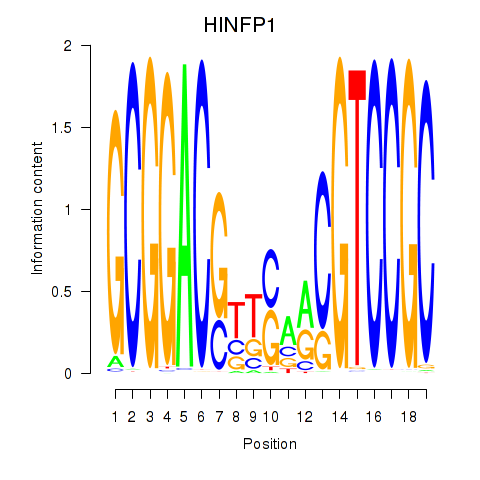
Results for HINFP1
Z-value: 0.36
Transcription factors associated with HINFP1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of HINFP1 motif
Sorted Z-values of HINFP1 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_61658853 | 0.76 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr11_-_61659006 | 0.61 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr18_+_12308231 | 0.50 |
ENST00000590103.1 ENST00000591909.1 ENST00000586653.1 ENST00000592683.1 ENST00000590967.1 ENST00000591208.1 ENST00000591463.1 |
TUBB6 |
tubulin, beta 6 class V |
| chr18_+_12308046 | 0.47 |
ENST00000317702.5 |
TUBB6 |
tubulin, beta 6 class V |
| chr1_-_94147385 | 0.29 |
ENST00000260502.6 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr6_-_56707943 | 0.28 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr9_-_33264676 | 0.27 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr3_+_133293278 | 0.26 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr3_+_133292759 | 0.18 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chrX_-_153707246 | 0.17 |
ENST00000407062.1 |
LAGE3 |
L antigen family, member 3 |
| chr2_-_234763147 | 0.17 |
ENST00000411486.2 ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP |
Holliday junction recognition protein |
| chr3_+_133292851 | 0.17 |
ENST00000503932.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr15_-_58357932 | 0.17 |
ENST00000347587.3 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr2_-_96811170 | 0.16 |
ENST00000288943.4 |
DUSP2 |
dual specificity phosphatase 2 |
| chr10_-_97453650 | 0.15 |
ENST00000371209.5 ENST00000371217.5 ENST00000430368.2 |
TCTN3 |
tectonic family member 3 |
| chr15_-_58357866 | 0.13 |
ENST00000537372.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr5_+_154393260 | 0.12 |
ENST00000435029.4 |
KIF4B |
kinesin family member 4B |
| chr1_+_43232913 | 0.11 |
ENST00000372525.5 ENST00000536543.1 |
C1orf50 |
chromosome 1 open reading frame 50 |
| chr18_+_11981547 | 0.11 |
ENST00000588927.1 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr14_+_73704201 | 0.11 |
ENST00000340738.5 ENST00000427855.1 ENST00000381166.3 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr6_-_144416737 | 0.10 |
ENST00000367569.2 |
SF3B5 |
splicing factor 3b, subunit 5, 10kDa |
| chr1_-_43232649 | 0.09 |
ENST00000372526.2 ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr20_-_3996165 | 0.09 |
ENST00000545616.2 ENST00000358395.6 |
RNF24 |
ring finger protein 24 |
| chr10_-_977564 | 0.08 |
ENST00000406525.2 |
LARP4B |
La ribonucleoprotein domain family, member 4B |
| chr17_-_79481666 | 0.08 |
ENST00000575659.1 |
ACTG1 |
actin, gamma 1 |
| chr16_+_57279248 | 0.08 |
ENST00000562023.1 ENST00000563234.1 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr10_-_75634219 | 0.07 |
ENST00000305762.7 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_-_75634260 | 0.05 |
ENST00000372765.1 ENST00000351293.3 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_-_75634326 | 0.05 |
ENST00000322635.3 ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_+_49514698 | 0.03 |
ENST00000432379.1 ENST00000429041.1 ENST00000374189.1 |
MAPK8 |
mitogen-activated protein kinase 8 |
| chr11_+_637246 | 0.02 |
ENST00000176183.5 |
DRD4 |
dopamine receptor D4 |
| chr17_-_2206801 | 0.02 |
ENST00000544865.1 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr21_+_35445827 | 0.01 |
ENST00000608209.1 ENST00000381151.3 |
SLC5A3 SLC5A3 |
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr19_-_14228541 | 0.01 |
ENST00000590853.1 ENST00000308677.4 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |