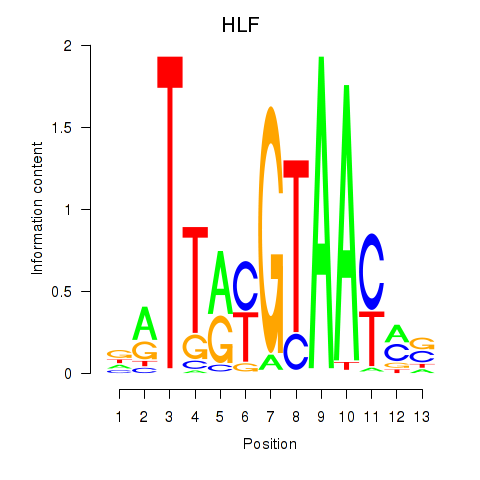
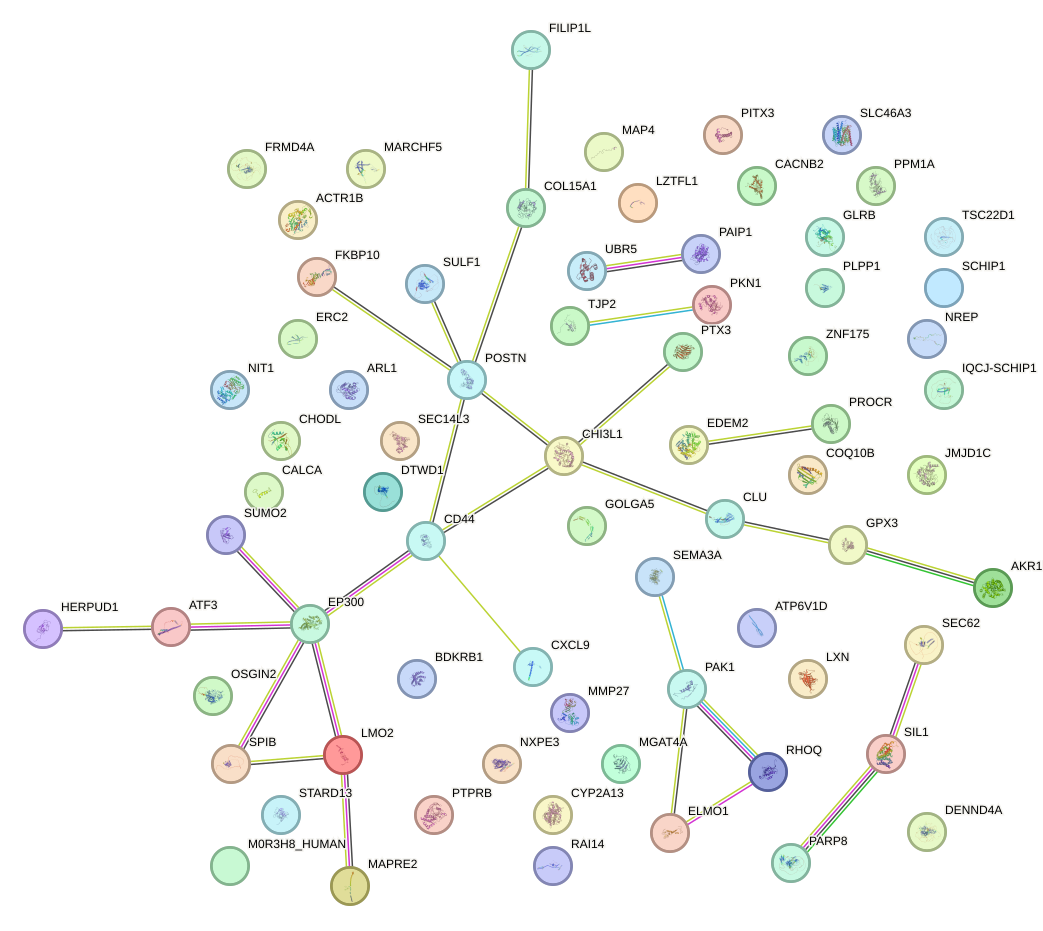
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HLF_TEF
Z-value: 0.57
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF |
|
TEF
|
ENSG00000167074.10 | TEF |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLF | hg19_v2_chr17_+_53342311_53342400 | -0.26 | 3.3e-01 | Click! |
| TEF | hg19_v2_chr22_+_41763274_41763337, hg19_v2_chr22_+_41777927_41777971 | -0.01 | 9.8e-01 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_158390282 | 2.78 |
ENST00000264265.3 |
LXN |
latexin |
| chr3_-_99594948 | 1.19 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr8_-_27469196 | 1.13 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr3_-_99595037 | 1.11 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr8_-_27468842 | 1.08 |
ENST00000523500.1 |
CLU |
clusterin |
| chr8_-_27468945 | 1.08 |
ENST00000405140.3 |
CLU |
clusterin |
| chr20_+_33759854 | 0.85 |
ENST00000216968.4 |
PROCR |
protein C receptor, endothelial |
| chr5_-_111091948 | 0.82 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr3_+_157154578 | 0.82 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr13_-_38172863 | 0.64 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr1_-_244013384 | 0.62 |
ENST00000366539.1 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr16_-_53537105 | 0.54 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr14_-_23285011 | 0.50 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_+_35201826 | 0.49 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr14_-_23285069 | 0.48 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_46459099 | 0.46 |
ENST00000371374.1 |
RCAN2 |
regulator of calcineurin 2 |
| chr3_-_48130707 | 0.44 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr10_-_14372870 | 0.42 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr3_+_158787041 | 0.41 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr20_-_33732952 | 0.40 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_-_71031185 | 0.39 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr9_+_71820057 | 0.37 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr9_+_71819927 | 0.36 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr3_-_48130314 | 0.36 |
ENST00000439356.1 ENST00000395734.3 ENST00000426837.2 |
MAP4 |
microtubule-associated protein 4 |
| chr21_+_27011584 | 0.35 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr3_+_62304712 | 0.34 |
ENST00000494481.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr1_+_212738676 | 0.33 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr5_-_54830871 | 0.33 |
ENST00000307259.8 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr3_+_62304648 | 0.31 |
ENST00000462069.1 ENST00000232519.5 ENST00000465142.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr5_+_34757309 | 0.30 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr2_-_203103185 | 0.29 |
ENST00000409205.1 |
SUMO1 |
small ubiquitin-like modifier 1 |
| chr2_+_46769798 | 0.27 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr3_+_101504200 | 0.26 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr8_+_70404996 | 0.26 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr3_+_154797877 | 0.26 |
ENST00000462745.1 ENST00000493237.1 |
MME |
membrane metallo-endopeptidase |
| chr17_-_34524157 | 0.26 |
ENST00000378354.4 ENST00000394484.1 |
CCL3L3 |
chemokine (C-C motif) ligand 3-like 3 |
| chr15_-_66084428 | 0.25 |
ENST00000443035.3 ENST00000431932.2 |
DENND4A |
DENN/MADD domain containing 4A |
| chr17_-_34625719 | 0.25 |
ENST00000422211.2 ENST00000542124.1 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
| chr14_+_93260569 | 0.24 |
ENST00000163416.2 |
GOLGA5 |
golgin A5 |
| chr12_-_71031220 | 0.24 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr14_+_93260642 | 0.22 |
ENST00000355976.2 |
GOLGA5 |
golgin A5 |
| chr7_-_37026108 | 0.21 |
ENST00000396045.3 |
ELMO1 |
engulfment and cell motility 1 |
| chr13_-_33780133 | 0.20 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr5_+_150404904 | 0.20 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr14_+_96722152 | 0.19 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr5_-_138534071 | 0.19 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr1_+_92632542 | 0.19 |
ENST00000409154.4 ENST00000370378.4 |
KIAA1107 |
KIAA1107 |
| chr2_+_191208196 | 0.18 |
ENST00000392329.2 ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1 |
inositol polyphosphate-1-phosphatase |
| chr9_-_88356789 | 0.18 |
ENST00000357081.3 ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1 |
ATP/GTP binding protein 1 |
| chr4_+_157997273 | 0.18 |
ENST00000541722.1 ENST00000512619.1 |
GLRB |
glycine receptor, beta |
| chr18_+_32621324 | 0.17 |
ENST00000300249.5 ENST00000538170.2 ENST00000588910.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr9_+_101705893 | 0.17 |
ENST00000375001.3 |
COL15A1 |
collagen, type XV, alpha 1 |
| chr2_-_99279928 | 0.16 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr4_-_76928641 | 0.15 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr7_-_83824169 | 0.15 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_+_52076425 | 0.15 |
ENST00000436511.2 |
ZNF175 |
zinc finger protein 175 |
| chr9_-_88356733 | 0.15 |
ENST00000376083.3 |
AGTPBP1 |
ATP/GTP binding protein 1 |
| chr5_-_138533973 | 0.14 |
ENST00000507002.1 ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr14_+_96722539 | 0.14 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr17_+_73089382 | 0.14 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_+_169079823 | 0.13 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr22_+_41487711 | 0.13 |
ENST00000263253.7 |
EP300 |
E1A binding protein p300 |
| chr13_-_29292956 | 0.13 |
ENST00000266943.6 |
SLC46A3 |
solute carrier family 46, member 3 |
| chr17_+_39975455 | 0.13 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr10_+_94050913 | 0.13 |
ENST00000358935.2 |
MARCH5 |
membrane-associated ring finger (C3HC4) 5 |
| chr19_+_49259325 | 0.12 |
ENST00000222157.3 |
FGF21 |
fibroblast growth factor 21 |
| chr19_+_50922187 | 0.12 |
ENST00000595883.1 ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr9_+_124103625 | 0.12 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr3_-_58200398 | 0.12 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr2_-_98280383 | 0.12 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr20_-_49547910 | 0.11 |
ENST00000396032.3 |
ADNP |
activity-dependent neuroprotector homeobox |
| chr19_+_49258775 | 0.11 |
ENST00000593756.1 |
FGF21 |
fibroblast growth factor 21 |
| chr2_+_198318147 | 0.11 |
ENST00000263960.2 |
COQ10B |
coenzyme Q10 homolog B (S. cerevisiae) |
| chr19_+_41594377 | 0.10 |
ENST00000330436.3 |
CYP2A13 |
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr1_-_203155868 | 0.10 |
ENST00000255409.3 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr14_+_60716159 | 0.09 |
ENST00000325658.3 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr8_-_103425047 | 0.09 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr11_-_77185094 | 0.08 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr13_-_45010939 | 0.08 |
ENST00000261489.2 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr5_+_49963239 | 0.08 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr10_+_18629628 | 0.08 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_-_67826486 | 0.07 |
ENST00000555431.1 ENST00000554236.1 ENST00000555474.1 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr5_+_95997918 | 0.07 |
ENST00000395812.2 ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST |
calpastatin |
| chr22_-_30867973 | 0.07 |
ENST00000402286.1 ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3 |
SEC14-like 3 (S. cerevisiae) |
| chr16_+_56965960 | 0.07 |
ENST00000439977.2 ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr7_+_100210133 | 0.07 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr5_+_95997769 | 0.07 |
ENST00000338252.3 ENST00000508830.1 |
CAST |
calpastatin |
| chr3_+_169684553 | 0.07 |
ENST00000337002.4 ENST00000480708.1 |
SEC62 |
SEC62 homolog (S. cerevisiae) |
| chr21_+_19617140 | 0.06 |
ENST00000299295.2 ENST00000338326.3 |
CHODL |
chondrolectin |
| chr20_-_49547731 | 0.06 |
ENST00000396029.3 |
ADNP |
activity-dependent neuroprotector homeobox |
| chr3_-_45883558 | 0.06 |
ENST00000445698.1 ENST00000296135.6 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr18_+_77160282 | 0.06 |
ENST00000318065.5 ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr10_-_65028938 | 0.06 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr11_-_14993819 | 0.06 |
ENST00000396372.2 ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA |
calcitonin-related polypeptide alpha |
| chr5_-_43557791 | 0.06 |
ENST00000338972.4 ENST00000511321.1 ENST00000515338.1 |
PAIP1 |
poly(A) binding protein interacting protein 1 |
| chr8_+_90914073 | 0.06 |
ENST00000297438.2 |
OSGIN2 |
oxidative stress induced growth inhibitor family member 2 |
| chr17_+_61851157 | 0.06 |
ENST00000578681.1 ENST00000583590.1 |
DDX42 |
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr1_+_161087873 | 0.05 |
ENST00000368009.2 ENST00000368007.4 ENST00000368008.1 ENST00000392190.5 |
NIT1 |
nitrilase 1 |
| chr13_-_46626847 | 0.05 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr15_+_49913201 | 0.05 |
ENST00000329873.5 ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1 |
DTW domain containing 1 |
| chr17_-_73179046 | 0.05 |
ENST00000314523.7 ENST00000420826.2 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr8_-_103424986 | 0.05 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr16_+_70332956 | 0.05 |
ENST00000288071.6 ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B RP11-529K1.3 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr12_-_101801505 | 0.05 |
ENST00000539055.1 ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1 |
ADP-ribosylation factor-like 1 |
| chr5_+_95998070 | 0.04 |
ENST00000421689.2 ENST00000510756.1 ENST00000512620.1 |
CAST |
calpastatin |
| chr11_-_33913708 | 0.04 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr11_-_102576537 | 0.04 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr5_-_147162263 | 0.04 |
ENST00000333010.6 ENST00000265272.5 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr17_-_73178599 | 0.04 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr10_-_65028817 | 0.04 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr3_+_88188254 | 0.03 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr7_+_44646177 | 0.03 |
ENST00000443864.2 ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr6_+_36238237 | 0.03 |
ENST00000457797.1 ENST00000394571.2 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
| chr8_-_103424916 | 0.03 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_-_37824442 | 0.03 |
ENST00000345060.3 |
ADRB3 |
adrenoceptor beta 3 |
| chr17_-_73851285 | 0.03 |
ENST00000589642.1 ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2 |
WW domain binding protein 2 |
| chr19_-_41945804 | 0.03 |
ENST00000221943.9 ENST00000597457.1 ENST00000589970.1 ENST00000595425.1 ENST00000438807.3 ENST00000589102.1 ENST00000592922.2 |
ATP5SL |
ATP5S-like |
| chr14_+_102276192 | 0.03 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_22259845 | 0.03 |
ENST00000420196.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr9_-_95298314 | 0.03 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr4_-_96470350 | 0.02 |
ENST00000504962.1 ENST00000453304.1 ENST00000506749.1 |
UNC5C |
unc-5 homolog C (C. elegans) |
| chr8_+_107738240 | 0.02 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr11_-_62313090 | 0.02 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr1_+_115572415 | 0.02 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr2_-_100106419 | 0.02 |
ENST00000393445.3 ENST00000258428.3 |
REV1 |
REV1, polymerase (DNA directed) |
| chr7_+_20686946 | 0.02 |
ENST00000443026.2 ENST00000406935.1 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_-_161207875 | 0.02 |
ENST00000512372.1 ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr2_-_167232484 | 0.02 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr8_+_24298531 | 0.02 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr18_+_3262954 | 0.02 |
ENST00000584539.1 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr17_-_41984835 | 0.02 |
ENST00000520406.1 ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr8_+_94752349 | 0.02 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr11_+_123986069 | 0.01 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr1_-_161208013 | 0.01 |
ENST00000515452.1 ENST00000367983.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr16_+_56899114 | 0.01 |
ENST00000566786.1 ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3 |
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr8_-_6735451 | 0.01 |
ENST00000297439.3 |
DEFB1 |
defensin, beta 1 |
| chr1_-_161207953 | 0.01 |
ENST00000367982.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr12_-_53074182 | 0.01 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr16_-_30102547 | 0.01 |
ENST00000279386.2 |
TBX6 |
T-box 6 |
| chr15_+_58430368 | 0.00 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr11_-_33795893 | 0.00 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chr22_-_29711704 | 0.00 |
ENST00000216101.6 |
RASL10A |
RAS-like, family 10, member A |
| chr1_-_161207986 | 0.00 |
ENST00000506209.1 ENST00000367980.2 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_186383741 | 0.00 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr10_-_97416400 | 0.00 |
ENST00000371224.2 ENST00000371221.3 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_116447283 | 0.00 |
ENST00000452729.1 ENST00000243222.4 |
COL10A1 |
collagen, type X, alpha 1 |
| chr14_-_70263979 | 0.00 |
ENST00000216540.4 |
SLC10A1 |
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 1.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 0.8 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.8 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.3 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.8 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |