Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
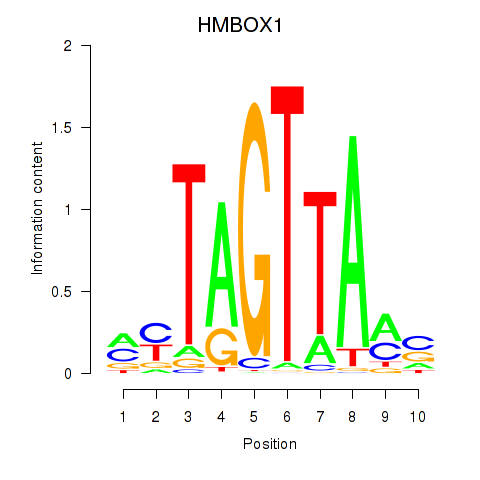
Results for HMBOX1
Z-value: 1.13
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.13 | HMBOX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg19_v2_chr8_+_28748099_28748142, hg19_v2_chr8_+_28748765_28748833 | -0.08 | 7.8e-01 | Click! |
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_149095652 | 4.40 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr1_-_173176452 | 2.54 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr10_+_89419370 | 2.53 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr11_+_19798964 | 2.11 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr8_+_30244580 | 2.05 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr7_-_47579188 | 2.01 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chrX_-_10851762 | 1.98 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr4_-_114900831 | 1.90 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr12_-_77272765 | 1.66 |
ENST00000547435.1 ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2 |
cysteine and glycine-rich protein 2 |
| chr7_-_120498357 | 1.60 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr12_+_75874580 | 1.54 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chrX_-_21776281 | 1.47 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr20_+_43211149 | 1.47 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chrX_-_10588459 | 1.41 |
ENST00000380782.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr5_+_92919043 | 1.40 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr1_-_110283138 | 1.39 |
ENST00000256594.3 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr7_+_134576317 | 1.39 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr17_-_3571934 | 1.38 |
ENST00000225525.3 |
TAX1BP3 |
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chrX_-_10588595 | 1.35 |
ENST00000423614.1 ENST00000317552.4 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr8_-_38326119 | 1.34 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr1_+_84609944 | 1.28 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_-_38326139 | 1.26 |
ENST00000335922.5 ENST00000532791.1 ENST00000397091.5 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chrX_+_66764375 | 1.26 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr9_+_75766652 | 1.25 |
ENST00000257497.6 |
ANXA1 |
annexin A1 |
| chr18_-_53253323 | 1.16 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chrX_+_37208540 | 1.14 |
ENST00000466533.1 ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1 TM4SF2 |
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr7_+_134576151 | 1.12 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr8_-_67525473 | 1.10 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chrX_+_37208521 | 1.09 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr4_-_186877502 | 1.08 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_-_201346761 | 0.95 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr1_-_152009460 | 0.90 |
ENST00000271638.2 |
S100A11 |
S100 calcium binding protein A11 |
| chr20_+_61584026 | 0.89 |
ENST00000370351.4 ENST00000370349.3 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr9_-_73029540 | 0.89 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr7_+_128379449 | 0.87 |
ENST00000479257.1 |
CALU |
calumenin |
| chr8_+_27491572 | 0.82 |
ENST00000301904.3 |
SCARA3 |
scavenger receptor class A, member 3 |
| chr19_+_16178317 | 0.77 |
ENST00000344824.6 ENST00000538887.1 |
TPM4 |
tropomyosin 4 |
| chr17_-_1395954 | 0.74 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chr14_+_45431379 | 0.68 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr5_-_77844974 | 0.63 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr13_-_67802549 | 0.62 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr4_-_110723194 | 0.61 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr12_+_81101277 | 0.58 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr22_-_37880543 | 0.56 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_52628271 | 0.55 |
ENST00000493422.1 |
GSTA2 |
glutathione S-transferase alpha 2 |
| chr4_-_110723134 | 0.52 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr22_-_37172111 | 0.52 |
ENST00000417951.2 ENST00000430701.1 ENST00000433985.2 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr6_-_165989936 | 0.52 |
ENST00000354448.4 |
PDE10A |
phosphodiesterase 10A |
| chr5_+_140762268 | 0.51 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr3_+_147127142 | 0.50 |
ENST00000282928.4 |
ZIC1 |
Zic family member 1 |
| chr22_-_37172191 | 0.49 |
ENST00000340630.5 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr5_+_140810132 | 0.48 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr6_+_46097711 | 0.47 |
ENST00000321037.4 |
ENPP4 |
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) |
| chr3_+_33155444 | 0.47 |
ENST00000320954.6 |
CRTAP |
cartilage associated protein |
| chr10_+_124320195 | 0.47 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr12_-_56236734 | 0.47 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr19_-_19051103 | 0.46 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr3_+_101504200 | 0.44 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr3_-_69101461 | 0.41 |
ENST00000543976.1 |
TMF1 |
TATA element modulatory factor 1 |
| chrX_+_70586140 | 0.41 |
ENST00000276072.3 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr12_+_54674482 | 0.41 |
ENST00000547708.1 ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
| chr17_-_10452929 | 0.41 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr10_-_112678976 | 0.40 |
ENST00000448814.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr6_-_52710893 | 0.40 |
ENST00000284562.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr2_+_162087577 | 0.39 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr1_+_87595433 | 0.38 |
ENST00000469312.2 ENST00000490006.2 |
RP5-1052I5.1 |
long intergenic non-protein coding RNA 1140 |
| chr11_-_59950519 | 0.37 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr11_-_59950622 | 0.37 |
ENST00000323961.3 ENST00000412309.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr1_+_198126093 | 0.36 |
ENST00000367385.4 ENST00000442588.1 ENST00000538004.1 |
NEK7 |
NIMA-related kinase 7 |
| chr10_-_112678904 | 0.36 |
ENST00000423273.1 ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr11_-_64825993 | 0.34 |
ENST00000340252.4 ENST00000355721.3 ENST00000356632.3 ENST00000355369.2 ENST00000339885.2 ENST00000358658.3 |
NAALADL1 |
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr14_-_50999307 | 0.34 |
ENST00000013125.4 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_+_33155525 | 0.34 |
ENST00000449224.1 |
CRTAP |
cartilage associated protein |
| chr2_+_24714729 | 0.33 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr6_-_52668605 | 0.32 |
ENST00000334575.5 |
GSTA1 |
glutathione S-transferase alpha 1 |
| chr3_-_69101413 | 0.32 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chrX_-_153523462 | 0.31 |
ENST00000361930.3 ENST00000369926.1 |
TEX28 |
testis expressed 28 |
| chr20_-_43133491 | 0.30 |
ENST00000411544.1 |
SERINC3 |
serine incorporator 3 |
| chr4_-_159080806 | 0.29 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr6_+_138725343 | 0.29 |
ENST00000607197.1 ENST00000367697.3 |
HEBP2 |
heme binding protein 2 |
| chr12_-_54673871 | 0.28 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr2_-_99952769 | 0.28 |
ENST00000409434.1 ENST00000434323.1 ENST00000264255.3 |
TXNDC9 |
thioredoxin domain containing 9 |
| chr2_+_27994567 | 0.27 |
ENST00000379666.3 ENST00000296102.3 |
MRPL33 |
mitochondrial ribosomal protein L33 |
| chr15_+_79166065 | 0.26 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr11_-_59950486 | 0.26 |
ENST00000426738.2 ENST00000533023.1 ENST00000420732.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr2_+_128848740 | 0.24 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr14_+_22615942 | 0.24 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr8_-_63951730 | 0.24 |
ENST00000260118.6 |
GGH |
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr2_-_220110111 | 0.23 |
ENST00000428427.1 ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L |
galactosidase, beta 1-like |
| chr3_+_69928256 | 0.23 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr11_+_64692143 | 0.23 |
ENST00000164133.2 ENST00000532850.1 |
PPP2R5B |
protein phosphatase 2, regulatory subunit B', beta |
| chr8_+_94929969 | 0.23 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr14_+_105331596 | 0.22 |
ENST00000556508.1 ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B |
centrosomal protein 170B |
| chr16_-_71842941 | 0.21 |
ENST00000423132.2 ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr2_+_204193149 | 0.21 |
ENST00000422511.2 |
ABI2 |
abl-interactor 2 |
| chr6_-_31940065 | 0.20 |
ENST00000375349.3 ENST00000337523.5 |
DXO |
decapping exoribonuclease |
| chr8_+_120885949 | 0.18 |
ENST00000523492.1 ENST00000286234.5 |
DEPTOR |
DEP domain containing MTOR-interacting protein |
| chrX_+_70586082 | 0.18 |
ENST00000373790.4 ENST00000449580.1 ENST00000423759.1 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr17_+_48172639 | 0.17 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr1_+_206643787 | 0.16 |
ENST00000367120.3 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr1_+_196743943 | 0.16 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr2_+_204193101 | 0.16 |
ENST00000430418.1 ENST00000424558.1 ENST00000261016.6 |
ABI2 |
abl-interactor 2 |
| chr12_+_72080253 | 0.16 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chr2_+_204193129 | 0.15 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr11_+_1860832 | 0.15 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr11_-_47447970 | 0.15 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr6_-_117747015 | 0.15 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr7_+_101460882 | 0.15 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr12_-_25348007 | 0.15 |
ENST00000354189.5 ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1 |
cancer susceptibility candidate 1 |
| chr16_+_28835437 | 0.15 |
ENST00000568266.1 |
ATXN2L |
ataxin 2-like |
| chr4_+_74301880 | 0.15 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr4_-_10117949 | 0.14 |
ENST00000508079.1 |
WDR1 |
WD repeat domain 1 |
| chr11_+_65769550 | 0.14 |
ENST00000312175.2 ENST00000445560.2 ENST00000530204.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr1_+_89246647 | 0.14 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chr16_-_71843047 | 0.14 |
ENST00000299980.4 ENST00000393512.3 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr3_-_45837959 | 0.13 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr13_-_77901177 | 0.13 |
ENST00000407578.2 ENST00000357337.6 ENST00000360084.5 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr2_-_220110187 | 0.13 |
ENST00000295759.7 ENST00000392089.2 |
GLB1L |
galactosidase, beta 1-like |
| chr15_+_75639296 | 0.13 |
ENST00000564500.1 ENST00000355059.4 ENST00000566752.1 |
NEIL1 |
nei endonuclease VIII-like 1 (E. coli) |
| chr1_-_169396666 | 0.12 |
ENST00000456107.1 ENST00000367805.3 |
CCDC181 |
coiled-coil domain containing 181 |
| chr1_-_169396646 | 0.12 |
ENST00000367806.3 |
CCDC181 |
coiled-coil domain containing 181 |
| chr8_-_101734170 | 0.12 |
ENST00000522387.1 ENST00000518196.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_176560742 | 0.12 |
ENST00000439151.2 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr4_+_156824840 | 0.11 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr19_+_42300548 | 0.11 |
ENST00000344550.4 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr19_+_55014085 | 0.11 |
ENST00000351841.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr5_+_176561129 | 0.10 |
ENST00000511258.1 ENST00000347982.4 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr3_-_45838011 | 0.10 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr19_+_7599128 | 0.09 |
ENST00000545201.2 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr1_-_150669500 | 0.09 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr4_-_69434245 | 0.09 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr19_+_55014013 | 0.09 |
ENST00000301202.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr3_-_49142504 | 0.08 |
ENST00000306125.6 ENST00000420147.2 |
QARS |
glutaminyl-tRNA synthetase |
| chr7_+_107220899 | 0.08 |
ENST00000379117.2 ENST00000473124.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr1_-_203274418 | 0.07 |
ENST00000457348.1 |
RP11-134P9.1 |
long intergenic non-protein coding RNA 1136 |
| chr17_-_3195876 | 0.07 |
ENST00000323404.1 |
OR3A1 |
olfactory receptor, family 3, subfamily A, member 1 |
| chr8_-_27630102 | 0.07 |
ENST00000356537.4 ENST00000522915.1 ENST00000539095.1 |
CCDC25 |
coiled-coil domain containing 25 |
| chr13_-_108870623 | 0.07 |
ENST00000405925.1 |
LIG4 |
ligase IV, DNA, ATP-dependent |
| chr12_+_117013656 | 0.06 |
ENST00000556529.1 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
| chr22_-_36635684 | 0.06 |
ENST00000358502.5 |
APOL2 |
apolipoprotein L, 2 |
| chr19_+_50353944 | 0.06 |
ENST00000594151.1 ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr14_-_89878369 | 0.06 |
ENST00000553840.1 ENST00000556916.1 |
FOXN3 |
forkhead box N3 |
| chr16_-_67281413 | 0.06 |
ENST00000258201.4 |
FHOD1 |
formin homology 2 domain containing 1 |
| chr1_+_27561104 | 0.06 |
ENST00000361771.3 |
WDTC1 |
WD and tetratricopeptide repeats 1 |
| chr1_+_207943667 | 0.05 |
ENST00000462968.2 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr1_+_196743912 | 0.05 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr19_+_38826477 | 0.05 |
ENST00000409410.2 ENST00000215069.4 |
CATSPERG |
catsper channel auxiliary subunit gamma |
| chr20_+_42187682 | 0.05 |
ENST00000373092.3 ENST00000373077.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr4_+_74347400 | 0.05 |
ENST00000226355.3 |
AFM |
afamin |
| chr19_+_38826415 | 0.04 |
ENST00000410018.1 ENST00000409235.3 |
CATSPERG |
catsper channel auxiliary subunit gamma |
| chr1_+_207226574 | 0.04 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr6_+_96969672 | 0.03 |
ENST00000369278.4 |
UFL1 |
UFM1-specific ligase 1 |
| chr22_+_24200085 | 0.03 |
ENST00000316185.8 |
SLC2A11 |
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr2_+_220110177 | 0.03 |
ENST00000409638.3 ENST00000396738.2 ENST00000409516.3 |
STK16 |
serine/threonine kinase 16 |
| chr10_+_112679301 | 0.03 |
ENST00000265277.5 ENST00000369452.4 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
| chr2_-_62081254 | 0.02 |
ENST00000405894.3 |
FAM161A |
family with sequence similarity 161, member A |
| chr11_-_62572901 | 0.02 |
ENST00000439713.2 ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1 |
nuclear RNA export factor 1 |
| chr14_+_56078695 | 0.01 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr6_+_134758827 | 0.01 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr1_-_86861936 | 0.01 |
ENST00000394733.2 ENST00000359242.3 ENST00000294678.2 ENST00000479890.1 ENST00000317336.7 ENST00000370567.1 ENST00000394731.1 ENST00000478286.2 ENST00000370566.3 |
ODF2L |
outer dense fiber of sperm tails 2-like |
| chr2_-_62081148 | 0.01 |
ENST00000404929.1 |
FAM161A |
family with sequence similarity 161, member A |
| chr14_-_23623577 | 0.00 |
ENST00000422941.2 ENST00000453702.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr12_-_57113333 | 0.00 |
ENST00000550920.1 |
NACA |
nascent polypeptide-associated complex alpha subunit |
| chr22_-_32334403 | 0.00 |
ENST00000543051.1 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr12_+_51633061 | 0.00 |
ENST00000551313.1 |
DAZAP2 |
DAZ associated protein 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 1.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 2.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 2.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 2.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 1.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.2 | GO:0001083 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 4.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.8 | 2.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 2.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 2.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.4 | 1.3 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.4 | 1.2 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.4 | 4.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 0.8 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.3 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.7 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.6 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 2.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.4 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 1.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.0 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 2.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.8 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.3 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 1.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.9 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.5 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.5 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.5 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.8 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |