Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
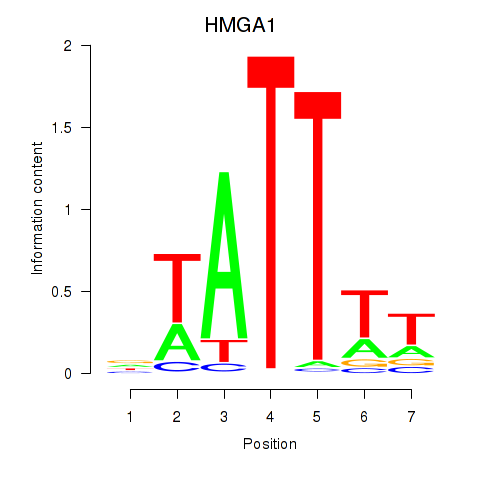
Results for HMGA1
Z-value: 0.62
Transcription factors associated with HMGA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA1
|
ENSG00000137309.15 | HMGA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA1 | hg19_v2_chr6_+_34204642_34204664 | 0.02 | 9.4e-01 | Click! |
Activity profile of HMGA1 motif
Sorted Z-values of HMGA1 motif
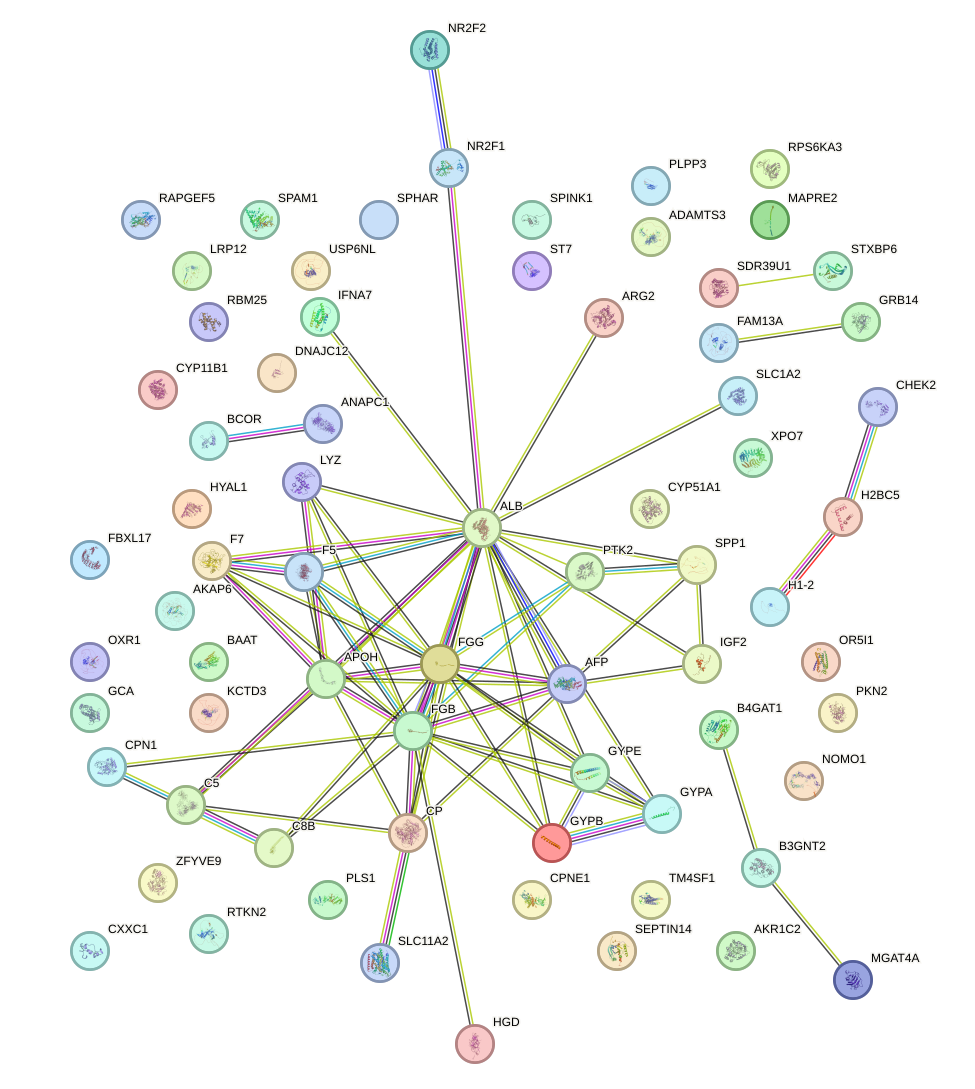
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74301880 | 3.78 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr4_+_155484155 | 1.81 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr5_+_92919043 | 1.65 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr4_+_74269956 | 1.37 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chrX_-_20236970 | 1.14 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr3_-_148939835 | 1.14 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr10_-_5046042 | 1.11 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr4_+_88896819 | 1.05 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr4_+_74275057 | 1.03 |
ENST00000511370.1 |
ALB |
albumin |
| chr4_-_155533787 | 0.99 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr4_-_110723194 | 0.94 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr4_-_110723134 | 0.93 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr17_-_64216748 | 0.92 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_+_69742121 | 0.90 |
ENST00000261267.2 ENST00000549690.1 ENST00000548839.1 |
LYZ |
lysozyme |
| chr17_-_64225508 | 0.89 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_100495864 | 0.87 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr4_-_110723335 | 0.76 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr11_+_22696314 | 0.75 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr10_+_114710516 | 0.74 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_-_22259845 | 0.67 |
ENST00000420196.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr3_-_120365866 | 0.67 |
ENST00000475447.2 |
HGD |
homogentisate 1,2-dioxygenase |
| chr4_+_155484103 | 0.65 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr10_-_69597915 | 0.64 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_-_99279928 | 0.62 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr6_+_153552455 | 0.58 |
ENST00000392385.2 |
AL590867.1 |
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr9_-_123812542 | 0.58 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr10_-_69597810 | 0.57 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_169555779 | 0.56 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr2_+_17721920 | 0.54 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr6_-_26056695 | 0.53 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr2_+_163175394 | 0.53 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr14_+_39736582 | 0.45 |
ENST00000556148.1 ENST00000348007.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr15_-_99789736 | 0.45 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr3_+_142342228 | 0.44 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr7_-_55930443 | 0.44 |
ENST00000388975.3 |
SEPT14 |
septin 14 |
| chr10_+_114710211 | 0.44 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr13_+_113760098 | 0.43 |
ENST00000346342.3 ENST00000541084.1 ENST00000375581.3 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr5_-_147211226 | 0.43 |
ENST00000296695.5 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr8_+_107738240 | 0.43 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr22_-_29107919 | 0.42 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr3_-_50340996 | 0.40 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr5_-_147211190 | 0.40 |
ENST00000510027.2 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr18_+_32556892 | 0.40 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_112614424 | 0.39 |
ENST00000427997.1 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr10_+_114709999 | 0.39 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_101825151 | 0.37 |
ENST00000441382.1 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr14_+_32798547 | 0.37 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr14_+_68086515 | 0.37 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr1_+_81771806 | 0.36 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr12_+_19358228 | 0.36 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr4_-_89744457 | 0.33 |
ENST00000395002.2 |
FAM13A |
family with sequence similarity 13, member A |
| chr2_-_165424973 | 0.32 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr9_-_21202204 | 0.32 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chrX_-_40036520 | 0.31 |
ENST00000406200.2 ENST00000378455.4 ENST00000342274.4 |
BCOR |
BCL6 corepressor |
| chr8_-_141931287 | 0.31 |
ENST00000517887.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr12_-_51402984 | 0.31 |
ENST00000545993.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_+_229440129 | 0.31 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr11_-_66115032 | 0.30 |
ENST00000311181.4 |
B3GNT1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr10_-_63995871 | 0.30 |
ENST00000315289.2 |
RTKN2 |
rhotekin 2 |
| chr10_+_114710425 | 0.30 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_+_22224811 | 0.29 |
ENST00000381237.1 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_91764108 | 0.28 |
ENST00000450723.1 |
CYP51A1 |
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr9_-_104145795 | 0.28 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr4_-_89744314 | 0.27 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr4_-_89744365 | 0.27 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr11_-_35440579 | 0.27 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_+_118812237 | 0.27 |
ENST00000513628.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr1_+_198608146 | 0.27 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr14_+_39703084 | 0.27 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr14_-_25479811 | 0.26 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr4_-_144940477 | 0.26 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr12_-_71551652 | 0.26 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr11_+_101983176 | 0.25 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr4_-_83765613 | 0.25 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr6_-_41039567 | 0.25 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr4_-_73434498 | 0.24 |
ENST00000286657.4 |
ADAMTS3 |
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr5_+_118812294 | 0.24 |
ENST00000509514.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chrX_+_9431324 | 0.24 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr18_+_32558208 | 0.24 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_196313239 | 0.24 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr11_-_55703876 | 0.23 |
ENST00000301532.3 |
OR5I1 |
olfactory receptor, family 5, subfamily I, member 1 |
| chr16_-_73093597 | 0.23 |
ENST00000397992.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr11_-_2162468 | 0.23 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_57431679 | 0.23 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr13_+_97928395 | 0.23 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr11_-_5255861 | 0.23 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr1_-_57045228 | 0.23 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr6_+_26158343 | 0.23 |
ENST00000377777.4 ENST00000289316.2 |
HIST1H2BD |
histone cluster 1, H2bd |
| chr3_+_109128837 | 0.22 |
ENST00000497996.1 |
RP11-702L6.4 |
RP11-702L6.4 |
| chr3_-_149095652 | 0.22 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr14_+_73563735 | 0.22 |
ENST00000532192.1 |
RBM25 |
RNA binding motif protein 25 |
| chr14_-_24911448 | 0.21 |
ENST00000555355.1 ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr16_+_14980632 | 0.21 |
ENST00000565655.1 |
NOMO1 |
NODAL modulator 1 |
| chr10_-_11574274 | 0.21 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr12_+_59989791 | 0.21 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_12524965 | 0.21 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_215740709 | 0.21 |
ENST00000259154.4 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
| chr20_+_11898507 | 0.21 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr1_+_89246647 | 0.20 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chr6_+_10556215 | 0.20 |
ENST00000316170.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr1_+_52682052 | 0.20 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr5_-_107703556 | 0.20 |
ENST00000496714.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr14_+_32798462 | 0.20 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr14_-_22005343 | 0.20 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr8_+_21823726 | 0.19 |
ENST00000433566.4 |
XPO7 |
exportin 7 |
| chr7_+_116654935 | 0.19 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr21_-_33975547 | 0.19 |
ENST00000431599.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr2_+_86333301 | 0.19 |
ENST00000254630.7 |
PTCD3 |
pentatricopeptide repeat domain 3 |
| chr3_+_124303539 | 0.19 |
ENST00000428018.2 |
KALRN |
kalirin, RhoGEF kinase |
| chr16_-_18908196 | 0.19 |
ENST00000565324.1 ENST00000561947.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_-_102591604 | 0.19 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr15_-_37390482 | 0.19 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr1_+_52082751 | 0.18 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr12_+_60083118 | 0.18 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr4_-_74088800 | 0.18 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr12_+_6602517 | 0.18 |
ENST00000315579.5 ENST00000539714.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr8_-_141774467 | 0.18 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr4_+_55095264 | 0.18 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_83769996 | 0.18 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr19_+_18208603 | 0.18 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr1_+_186265399 | 0.17 |
ENST00000367486.3 ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4 |
proteoglycan 4 |
| chr3_-_178969403 | 0.17 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_+_171561127 | 0.17 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chrX_+_37639302 | 0.17 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr5_-_98262240 | 0.17 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr14_-_90085458 | 0.17 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr12_+_41831485 | 0.17 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr5_+_140213815 | 0.16 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr1_-_220220000 | 0.16 |
ENST00000366923.3 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chrX_-_135962876 | 0.16 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr11_-_46615498 | 0.16 |
ENST00000533727.1 ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
| chr7_+_123488124 | 0.16 |
ENST00000476325.1 |
HYAL4 |
hyaluronoglucosaminidase 4 |
| chr18_+_47901365 | 0.16 |
ENST00000285116.3 |
SKA1 |
spindle and kinetochore associated complex subunit 1 |
| chr5_-_131347583 | 0.16 |
ENST00000379255.1 ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr6_+_26156551 | 0.16 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr4_+_39460689 | 0.16 |
ENST00000381846.1 |
LIAS |
lipoic acid synthetase |
| chr8_-_17555164 | 0.16 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr7_-_152373216 | 0.15 |
ENST00000359321.1 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr6_-_25830785 | 0.15 |
ENST00000468082.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chr17_+_37856299 | 0.15 |
ENST00000269571.5 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr3_+_73110810 | 0.15 |
ENST00000533473.1 |
EBLN2 |
endogenous Bornavirus-like nucleoprotein 2 |
| chr14_+_102276209 | 0.15 |
ENST00000445439.3 ENST00000334743.5 ENST00000557095.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_-_34420259 | 0.15 |
ENST00000250457.3 ENST00000547327.2 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
| chr9_+_706842 | 0.14 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr2_+_120687335 | 0.14 |
ENST00000544261.1 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr1_-_150669604 | 0.14 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr3_-_150996239 | 0.14 |
ENST00000309170.3 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
| chr16_+_72142195 | 0.14 |
ENST00000563819.1 ENST00000567142.2 |
DHX38 |
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr8_-_95274536 | 0.14 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr13_+_33160553 | 0.14 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_+_148508845 | 0.14 |
ENST00000491148.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr11_+_89867681 | 0.14 |
ENST00000534061.1 |
NAALAD2 |
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr11_+_93479588 | 0.14 |
ENST00000526335.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
| chr12_+_8995832 | 0.13 |
ENST00000541459.1 |
A2ML1 |
alpha-2-macroglobulin-like 1 |
| chr20_-_14318248 | 0.13 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr5_+_140207536 | 0.13 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr1_+_44870866 | 0.13 |
ENST00000355387.2 ENST00000361799.2 |
RNF220 |
ring finger protein 220 |
| chr6_+_20534672 | 0.13 |
ENST00000274695.4 ENST00000378624.4 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
| chr16_-_71610985 | 0.13 |
ENST00000355962.4 |
TAT |
tyrosine aminotransferase |
| chr4_+_79567314 | 0.13 |
ENST00000503539.1 ENST00000504675.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr9_-_70465758 | 0.13 |
ENST00000489273.1 |
CBWD5 |
COBW domain containing 5 |
| chr5_+_137203557 | 0.13 |
ENST00000515645.1 |
MYOT |
myotilin |
| chr10_+_32856764 | 0.13 |
ENST00000375030.2 ENST00000375028.3 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chrX_-_13835461 | 0.13 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr11_+_71249071 | 0.13 |
ENST00000398534.3 |
KRTAP5-8 |
keratin associated protein 5-8 |
| chr2_-_24270217 | 0.13 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chr5_+_137203541 | 0.12 |
ENST00000421631.2 |
MYOT |
myotilin |
| chr10_+_51572408 | 0.12 |
ENST00000374082.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr22_+_40573921 | 0.12 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr7_-_140482926 | 0.12 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr3_-_167191814 | 0.12 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr2_-_136678123 | 0.12 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr1_-_160231451 | 0.12 |
ENST00000495887.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr10_-_94003003 | 0.12 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr6_-_39197226 | 0.12 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr21_-_43735628 | 0.12 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr6_-_151773232 | 0.12 |
ENST00000444024.1 ENST00000367303.4 |
RMND1 |
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr14_-_21994525 | 0.12 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr4_-_153303658 | 0.12 |
ENST00000296555.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_-_22063787 | 0.12 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_67632083 | 0.11 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chr2_+_90198535 | 0.11 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr7_+_106505696 | 0.11 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr13_+_109248500 | 0.11 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr1_+_215747118 | 0.11 |
ENST00000448333.1 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
| chr14_+_102276132 | 0.11 |
ENST00000350249.3 ENST00000557621.1 ENST00000556946.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_-_130970723 | 0.11 |
ENST00000308008.6 ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr20_-_33999766 | 0.11 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr5_-_27038683 | 0.11 |
ENST00000511822.1 ENST00000231021.4 |
CDH9 |
cadherin 9, type 2 (T1-cadherin) |
| chr17_+_3379284 | 0.11 |
ENST00000263080.2 |
ASPA |
aspartoacylase |
| chr5_+_140501581 | 0.11 |
ENST00000194152.1 |
PCDHB4 |
protocadherin beta 4 |
| chr6_-_64029879 | 0.11 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr3_+_89156799 | 0.11 |
ENST00000452448.2 ENST00000494014.1 |
EPHA3 |
EPH receptor A3 |
| chr3_+_138340067 | 0.11 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr13_+_113633620 | 0.11 |
ENST00000421756.1 ENST00000375601.3 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr6_+_158733692 | 0.11 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr14_+_56078695 | 0.11 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr1_-_28384598 | 0.11 |
ENST00000373864.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr13_+_49684445 | 0.11 |
ENST00000398316.3 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr14_-_23504432 | 0.11 |
ENST00000425762.2 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.5 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 3.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 4.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.3 | 1.0 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 1.9 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 3.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 2.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 2.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 1.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.4 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.8 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.4 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.2 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.1 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 1.0 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.9 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0071283 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) cellular response to iron(III) ion(GO:0071283) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 2.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.0 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |