Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
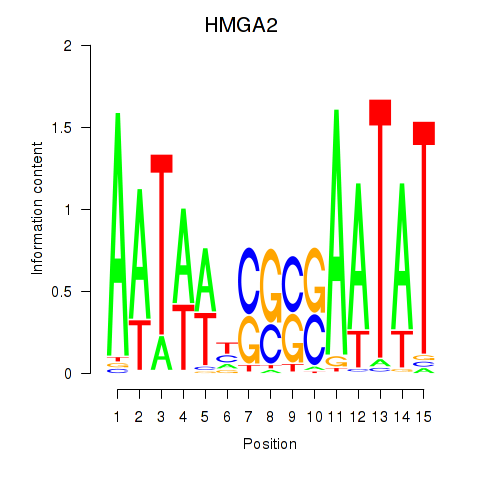
Results for HMGA2
Z-value: 0.63
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | HMGA2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218598_66218678 | 0.79 | 2.7e-04 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_5046042 | 2.35 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr15_+_58702742 | 2.35 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr14_-_65409438 | 1.84 |
ENST00000557049.1 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr14_-_65409502 | 1.61 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr2_-_216257849 | 1.46 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chrY_+_22918021 | 1.22 |
ENST00000288666.5 |
RPS4Y2 |
ribosomal protein S4, Y-linked 2 |
| chr19_-_36297632 | 1.18 |
ENST00000588266.2 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr3_+_193853927 | 1.02 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr14_-_94759595 | 1.00 |
ENST00000261994.4 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759361 | 0.98 |
ENST00000393096.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759408 | 0.97 |
ENST00000554723.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_235887329 | 0.79 |
ENST00000409212.1 ENST00000344528.4 ENST00000444916.1 |
SH3BP4 |
SH3-domain binding protein 4 |
| chr11_+_69455855 | 0.71 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr15_+_84904525 | 0.68 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr9_-_115480303 | 0.51 |
ENST00000374234.1 ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP |
INTS3 and NABP interacting protein |
| chr6_-_52705641 | 0.50 |
ENST00000370989.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr6_+_123110302 | 0.44 |
ENST00000368440.4 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr20_+_48884002 | 0.44 |
ENST00000425497.1 ENST00000445003.1 |
RP11-290F20.3 |
RP11-290F20.3 |
| chr2_+_220143989 | 0.41 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_+_120436733 | 0.39 |
ENST00000401466.1 ENST00000424086.1 ENST00000272521.6 |
TMEM177 |
transmembrane protein 177 |
| chr19_-_12267524 | 0.38 |
ENST00000455799.1 ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625 |
zinc finger protein 625 |
| chr11_+_73661364 | 0.37 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr6_+_111580508 | 0.36 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chr1_+_55464600 | 0.36 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr2_+_220144052 | 0.34 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr15_+_83098710 | 0.30 |
ENST00000561062.1 ENST00000358583.3 |
GOLGA6L9 |
golgin A6 family-like 20 |
| chr3_-_106959424 | 0.27 |
ENST00000607801.1 ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882 |
long intergenic non-protein coding RNA 882 |
| chr15_+_82722225 | 0.27 |
ENST00000300515.8 |
GOLGA6L9 |
golgin A6 family-like 9 |
| chr18_-_21017817 | 0.26 |
ENST00000542162.1 ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241 |
transmembrane protein 241 |
| chr12_+_21207503 | 0.26 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr17_-_4607335 | 0.25 |
ENST00000570571.1 ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1 |
proline, glutamate and leucine rich protein 1 |
| chr15_+_43985084 | 0.24 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr17_-_48785216 | 0.22 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr12_-_122751002 | 0.22 |
ENST00000267199.4 |
VPS33A |
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr5_+_150827143 | 0.22 |
ENST00000243389.3 ENST00000517945.1 ENST00000521925.1 |
SLC36A1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr15_+_43885252 | 0.22 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr20_+_46130619 | 0.21 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr20_+_46130601 | 0.20 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr4_+_40058411 | 0.19 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr14_+_45553296 | 0.19 |
ENST00000355765.6 ENST00000553605.1 |
PRPF39 |
pre-mRNA processing factor 39 |
| chr18_+_11851383 | 0.16 |
ENST00000526991.2 |
CHMP1B |
charged multivesicular body protein 1B |
| chr20_+_46130671 | 0.16 |
ENST00000371998.3 ENST00000371997.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr6_-_52926539 | 0.16 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr3_+_186501336 | 0.16 |
ENST00000323963.5 ENST00000440191.2 ENST00000356531.5 |
EIF4A2 |
eukaryotic translation initiation factor 4A2 |
| chr1_-_226374373 | 0.15 |
ENST00000366812.5 |
ACBD3 |
acyl-CoA binding domain containing 3 |
| chr2_-_89266286 | 0.14 |
ENST00000464162.1 |
IGKV1-6 |
immunoglobulin kappa variable 1-6 |
| chr19_-_38146289 | 0.13 |
ENST00000392144.1 ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30 |
ZFP30 zinc finger protein |
| chr20_-_31989307 | 0.13 |
ENST00000473997.1 ENST00000544843.1 ENST00000346416.2 ENST00000357886.4 ENST00000339269.5 |
CDK5RAP1 |
CDK5 regulatory subunit associated protein 1 |
| chr9_+_42704004 | 0.11 |
ENST00000457288.1 |
CBWD7 |
COBW domain containing 7 |
| chr7_-_102985035 | 0.11 |
ENST00000426036.2 ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2 |
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr12_+_18414446 | 0.10 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr7_-_102985288 | 0.07 |
ENST00000379263.3 |
DNAJC2 |
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chrX_-_135962876 | 0.06 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chrX_-_77150985 | 0.05 |
ENST00000358075.6 |
MAGT1 |
magnesium transporter 1 |
| chr13_+_50656307 | 0.05 |
ENST00000378180.4 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr11_+_67250490 | 0.04 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr3_+_137728842 | 0.04 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr9_+_108463234 | 0.03 |
ENST00000374688.1 |
TMEM38B |
transmembrane protein 38B |
| chr12_-_10324716 | 0.02 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr19_+_44455368 | 0.02 |
ENST00000591168.1 ENST00000587682.1 ENST00000251269.5 |
ZNF221 |
zinc finger protein 221 |
| chr17_-_50236039 | 0.02 |
ENST00000451037.2 |
CA10 |
carbonic anhydrase X |
| chr12_-_10573149 | 0.01 |
ENST00000381904.2 ENST00000381903.2 ENST00000396439.2 |
KLRC3 |
killer cell lectin-like receptor subfamily C, member 3 |
| chr15_+_28623784 | 0.01 |
ENST00000526619.2 ENST00000337838.7 ENST00000532622.2 |
GOLGA8F |
golgin A8 family, member F |
| chr5_+_36608422 | 0.00 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.4 | 3.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.0 | GO:0021558 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 2.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 1.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 2.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.6 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.7 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 3.2 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.8 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.4 | GO:0035082 | axoneme assembly(GO:0035082) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.3 | 1.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 3.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 3.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |