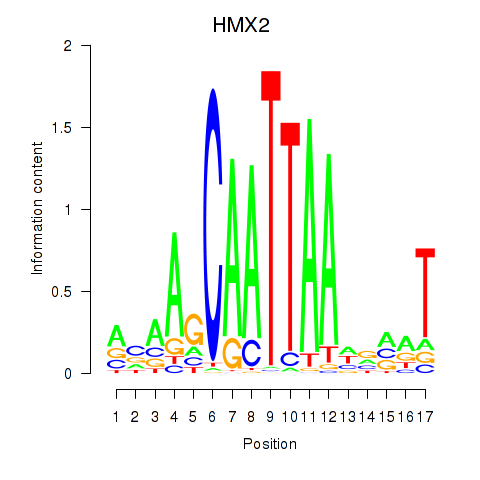
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HMX2
Z-value: 0.84
Transcription factors associated with HMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX2
|
ENSG00000188816.3 | HMX2 |
Activity profile of HMX2 motif
Sorted Z-values of HMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95241951 | 6.69 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr2_-_151344172 | 4.38 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr10_-_95242044 | 3.42 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr1_+_99127225 | 2.61 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr14_+_55595762 | 2.05 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr10_-_126849588 | 2.05 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr7_-_87856303 | 1.87 |
ENST00000394641.3 |
SRI |
sorcin |
| chr11_+_5710919 | 1.86 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr7_-_87856280 | 1.76 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr6_+_12290586 | 1.51 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr14_+_55595960 | 1.46 |
ENST00000554715.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr2_-_190044480 | 1.46 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_-_110933611 | 1.45 |
ENST00000472422.2 ENST00000437429.2 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr6_+_151561085 | 1.45 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr1_+_186798073 | 1.40 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr13_-_38172863 | 1.28 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr2_-_169104651 | 1.14 |
ENST00000355999.4 |
STK39 |
serine threonine kinase 39 |
| chr4_+_90823130 | 1.04 |
ENST00000508372.1 |
MMRN1 |
multimerin 1 |
| chr6_+_148663729 | 1.01 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr8_-_13134045 | 0.98 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr2_-_160654745 | 0.91 |
ENST00000259053.4 ENST00000429078.2 |
CD302 |
CD302 molecule |
| chr1_-_110933663 | 0.91 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr4_-_90759440 | 0.90 |
ENST00000336904.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_+_114178512 | 0.90 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr5_-_146781153 | 0.80 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr17_-_46671323 | 0.77 |
ENST00000239151.5 |
HOXB5 |
homeobox B5 |
| chr12_+_104680659 | 0.75 |
ENST00000526691.1 ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1 |
thioredoxin reductase 1 |
| chr5_+_119867159 | 0.75 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr3_+_122785895 | 0.74 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr5_+_140868717 | 0.73 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr1_+_144151520 | 0.70 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr3_-_79816965 | 0.64 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_-_205290865 | 0.63 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr6_-_46293378 | 0.61 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr8_+_32579341 | 0.52 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr2_-_20251744 | 0.50 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr10_-_101945771 | 0.49 |
ENST00000370408.2 ENST00000407654.3 |
ERLIN1 |
ER lipid raft associated 1 |
| chr19_+_40877583 | 0.49 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr2_+_24272543 | 0.49 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr2_+_152214098 | 0.49 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_111334678 | 0.45 |
ENST00000329516.3 ENST00000330331.5 ENST00000446930.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr4_+_166248775 | 0.44 |
ENST00000261507.6 ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1 |
methylsterol monooxygenase 1 |
| chr2_-_152590982 | 0.44 |
ENST00000409198.1 ENST00000397345.3 ENST00000427231.2 |
NEB |
nebulin |
| chr1_-_217804377 | 0.43 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr13_+_21714711 | 0.42 |
ENST00000607003.1 ENST00000492245.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr11_+_60681346 | 0.40 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr3_-_187009646 | 0.40 |
ENST00000296280.6 ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr11_-_327537 | 0.40 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr17_+_1936687 | 0.39 |
ENST00000570477.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr4_+_83956237 | 0.38 |
ENST00000264389.2 |
COPS4 |
COP9 signalosome subunit 4 |
| chr1_+_209878182 | 0.34 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr18_+_76829385 | 0.34 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chr4_+_83956312 | 0.32 |
ENST00000509317.1 ENST00000503682.1 ENST00000511653.1 |
COPS4 |
COP9 signalosome subunit 4 |
| chr1_-_21377447 | 0.32 |
ENST00000374937.3 ENST00000264211.8 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr5_+_140227048 | 0.32 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr10_-_121296045 | 0.31 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr2_-_55237484 | 0.31 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr12_+_133614119 | 0.30 |
ENST00000327668.7 |
ZNF84 |
zinc finger protein 84 |
| chr7_+_5920429 | 0.28 |
ENST00000242104.5 |
OCM |
oncomodulin |
| chr8_+_31496809 | 0.28 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr22_+_51176624 | 0.28 |
ENST00000216139.5 ENST00000529621.1 |
ACR |
acrosin |
| chr3_-_46249878 | 0.27 |
ENST00000296140.3 |
CCR1 |
chemokine (C-C motif) receptor 1 |
| chr13_-_103426112 | 0.27 |
ENST00000376032.4 ENST00000376029.3 |
TEX30 |
testis expressed 30 |
| chr3_-_114343039 | 0.26 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr9_+_27109133 | 0.26 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr4_-_48782259 | 0.25 |
ENST00000507711.1 ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL |
FRY-like |
| chr1_-_21377383 | 0.25 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr19_+_41281416 | 0.25 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr19_+_18668572 | 0.24 |
ENST00000540691.1 ENST00000539106.1 ENST00000222307.4 |
KXD1 |
KxDL motif containing 1 |
| chr13_-_103426081 | 0.23 |
ENST00000376022.1 ENST00000376021.4 |
TEX30 |
testis expressed 30 |
| chr7_+_141695633 | 0.23 |
ENST00000549489.2 |
MGAM |
maltase-glucoamylase (alpha-glucosidase) |
| chr15_-_55700457 | 0.23 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr10_-_49813090 | 0.22 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr3_+_111697843 | 0.22 |
ENST00000534857.1 ENST00000273359.3 ENST00000494817.1 |
ABHD10 |
abhydrolase domain containing 10 |
| chr1_+_115397424 | 0.22 |
ENST00000369522.3 ENST00000455987.1 |
SYCP1 |
synaptonemal complex protein 1 |
| chr19_+_18668672 | 0.21 |
ENST00000601630.1 ENST00000599000.1 ENST00000595073.1 ENST00000596785.1 |
KXD1 |
KxDL motif containing 1 |
| chr2_+_109237717 | 0.21 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr4_+_83821835 | 0.20 |
ENST00000302236.5 |
THAP9 |
THAP domain containing 9 |
| chr1_-_202858227 | 0.20 |
ENST00000367262.3 |
RABIF |
RAB interacting factor |
| chr7_+_141695671 | 0.20 |
ENST00000497673.1 ENST00000475668.2 |
MGAM |
maltase-glucoamylase (alpha-glucosidase) |
| chr5_+_167913450 | 0.20 |
ENST00000231572.3 ENST00000538719.1 |
RARS |
arginyl-tRNA synthetase |
| chr2_-_145277569 | 0.19 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr17_+_45973516 | 0.19 |
ENST00000376741.4 |
SP2 |
Sp2 transcription factor |
| chr4_-_122854612 | 0.19 |
ENST00000264811.5 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr17_+_8191815 | 0.18 |
ENST00000226105.6 ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF |
RAN guanine nucleotide release factor |
| chr4_-_159080806 | 0.18 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr19_-_51340469 | 0.18 |
ENST00000326856.4 |
KLK15 |
kallikrein-related peptidase 15 |
| chr1_-_213189168 | 0.17 |
ENST00000366962.3 ENST00000360506.2 |
ANGEL2 |
angel homolog 2 (Drosophila) |
| chr1_-_67266939 | 0.16 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr12_+_26164645 | 0.16 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr3_-_108836977 | 0.15 |
ENST00000232603.5 |
MORC1 |
MORC family CW-type zinc finger 1 |
| chr3_-_52713729 | 0.15 |
ENST00000296302.7 ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1 |
polybromo 1 |
| chr6_-_106773491 | 0.15 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr3_+_173116225 | 0.15 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr6_+_25754927 | 0.14 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr18_+_42260861 | 0.14 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr13_+_27998681 | 0.14 |
ENST00000381140.4 |
GTF3A |
general transcription factor IIIA |
| chr15_-_55700522 | 0.13 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr4_-_70518941 | 0.12 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr7_+_77325738 | 0.12 |
ENST00000334955.8 |
RSBN1L |
round spermatid basic protein 1-like |
| chr2_-_224467093 | 0.11 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr4_+_76649797 | 0.10 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr13_+_21714913 | 0.10 |
ENST00000450573.1 ENST00000467636.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr11_+_93517393 | 0.09 |
ENST00000251871.3 ENST00000530819.1 |
MED17 |
mediator complex subunit 17 |
| chrX_+_119737806 | 0.09 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chrX_+_1710484 | 0.09 |
ENST00000313871.3 ENST00000381261.3 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
| chr5_+_156607829 | 0.09 |
ENST00000422843.3 |
ITK |
IL2-inducible T-cell kinase |
| chr12_-_123450986 | 0.09 |
ENST00000344275.7 ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr11_-_104893863 | 0.08 |
ENST00000260315.3 ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5 |
caspase 5, apoptosis-related cysteine peptidase |
| chr5_-_142814241 | 0.08 |
ENST00000504572.1 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr19_+_9361606 | 0.08 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr16_-_21663919 | 0.07 |
ENST00000569602.1 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr22_-_39268308 | 0.07 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr2_+_27255806 | 0.06 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr2_+_1417228 | 0.06 |
ENST00000382269.3 ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO |
thyroid peroxidase |
| chr9_-_95055956 | 0.06 |
ENST00000375629.3 ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS |
isoleucyl-tRNA synthetase |
| chr19_+_54641444 | 0.06 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_216974020 | 0.06 |
ENST00000392132.2 ENST00000417391.1 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr3_-_194119995 | 0.06 |
ENST00000323007.3 |
GP5 |
glycoprotein V (platelet) |
| chr3_-_187009798 | 0.06 |
ENST00000337774.5 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr20_+_33104199 | 0.06 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr2_+_106468204 | 0.04 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr12_-_10151773 | 0.04 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr5_+_95066823 | 0.04 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr19_+_58144529 | 0.03 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr20_+_48429356 | 0.03 |
ENST00000361573.2 ENST00000541138.1 ENST00000539601.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr20_+_3776371 | 0.03 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr1_+_196946664 | 0.03 |
ENST00000367414.5 |
CFHR5 |
complement factor H-related 5 |
| chr5_-_22853429 | 0.02 |
ENST00000504376.2 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
| chr17_-_73663245 | 0.02 |
ENST00000584999.1 ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5 |
RecQ protein-like 5 |
| chrY_+_20137667 | 0.02 |
ENST00000250838.4 ENST00000426790.1 |
CDY2A |
chromodomain protein, Y-linked, 2A |
| chr22_+_39795746 | 0.02 |
ENST00000216160.6 ENST00000331454.3 |
TAB1 |
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr1_+_196946680 | 0.02 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr3_-_121379739 | 0.01 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr11_-_64684672 | 0.01 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr4_-_100212132 | 0.01 |
ENST00000209668.2 |
ADH1A |
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chrY_-_19992098 | 0.01 |
ENST00000544303.1 ENST00000382867.3 |
CDY2B |
chromodomain protein, Y-linked, 2B |
| chr7_-_14880892 | 0.01 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr11_+_122753391 | 0.00 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
| chr9_+_27109392 | 0.00 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.7 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.2 | 0.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0016160 | amylase activity(GO:0016160) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 3.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 10.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 10.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 3.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.8 | 10.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 3.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.5 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 1.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.4 | 1.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.4 | 1.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.4 | 1.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 2.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.6 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.9 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.2 | GO:0060259 | positive regulation of behavior(GO:0048520) regulation of feeding behavior(GO:0060259) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 1.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0098923 | NMDA glutamate receptor clustering(GO:0097114) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 1.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0070269 | ectopic germ cell programmed cell death(GO:0035234) pyroptosis(GO:0070269) |
| 0.0 | 2.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 10.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |