Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HMX3
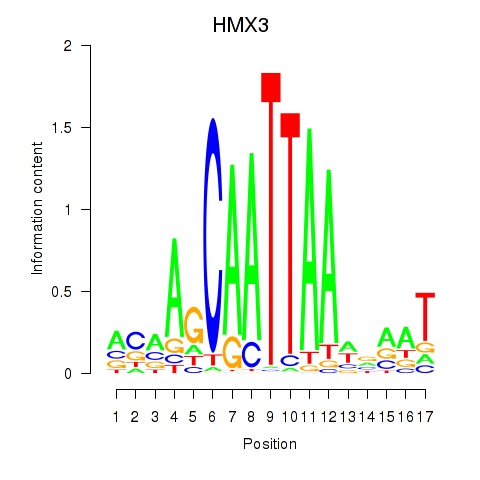
Z-value: 1.20
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | HMX3 |
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
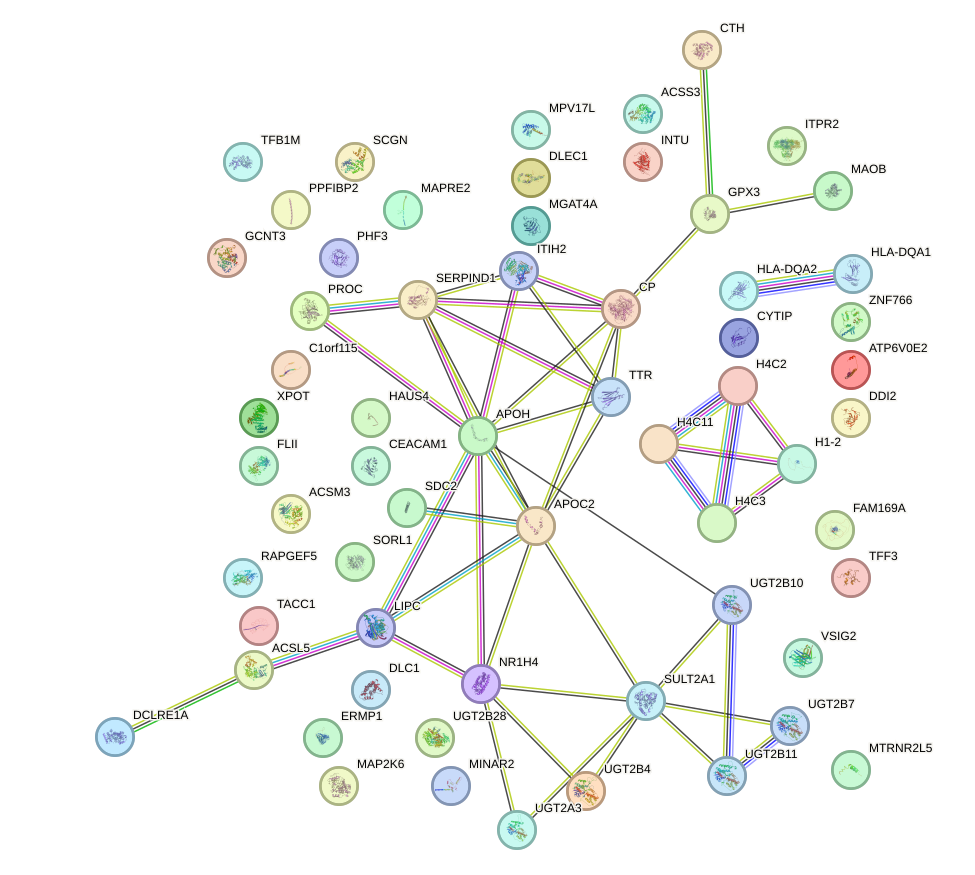
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_64225508 | 4.94 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_+_186330712 | 4.14 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr4_+_69681710 | 3.64 |
ENST00000265403.7 ENST00000458688.2 |
UGT2B10 |
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr4_+_69962212 | 2.90 |
ENST00000508661.1 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962185 | 2.90 |
ENST00000305231.7 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr5_+_150404904 | 2.74 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr2_-_99279928 | 2.28 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr18_+_29171689 | 2.26 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr22_+_21133469 | 2.22 |
ENST00000406799.1 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr9_+_103947311 | 2.16 |
ENST00000395056.2 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr4_-_155511887 | 2.07 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr4_-_69817481 | 2.02 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr12_+_51318513 | 1.91 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr18_+_32556892 | 1.86 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr15_+_58702742 | 1.80 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr12_+_100867486 | 1.73 |
ENST00000548884.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr19_-_48389651 | 1.72 |
ENST00000222002.3 |
SULT2A1 |
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr8_-_17752912 | 1.71 |
ENST00000398054.1 ENST00000381840.2 |
FGL1 |
fibrinogen-like 1 |
| chr10_+_7745303 | 1.70 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr19_+_45449301 | 1.60 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr8_-_17752996 | 1.59 |
ENST00000381841.2 ENST00000427924.1 |
FGL1 |
fibrinogen-like 1 |
| chr3_-_148939835 | 1.58 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr10_+_114135952 | 1.56 |
ENST00000356116.1 ENST00000433418.1 ENST00000354273.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr14_-_20801427 | 1.44 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr4_+_70146217 | 1.42 |
ENST00000335568.5 ENST00000511240.1 |
UGT2B28 |
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr3_+_169629354 | 1.38 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
| chr11_+_62104897 | 1.38 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr5_+_129083772 | 1.37 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr12_+_81471816 | 1.30 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr8_-_101571964 | 1.28 |
ENST00000520552.1 ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr4_-_110723194 | 1.26 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr4_-_110723134 | 1.26 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr1_+_220863187 | 1.24 |
ENST00000294889.5 |
C1orf115 |
chromosome 1 open reading frame 115 |
| chr12_+_100867694 | 1.23 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_87342564 | 1.19 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr8_-_101571933 | 1.19 |
ENST00000520311.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr8_+_95565947 | 1.16 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr7_-_22234381 | 1.14 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_70080449 | 1.11 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr12_-_26986076 | 1.10 |
ENST00000381340.3 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr19_+_52772821 | 1.10 |
ENST00000439461.1 |
ZNF766 |
zinc finger protein 766 |
| chr18_-_10701979 | 1.06 |
ENST00000538948.1 ENST00000285141.4 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr1_+_111415757 | 1.04 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr21_-_34185944 | 0.99 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr16_+_20775358 | 0.99 |
ENST00000440284.2 |
ACSM3 |
acyl-CoA synthetase medium-chain family member 3 |
| chr16_-_28634874 | 0.98 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr5_-_74162605 | 0.95 |
ENST00000389156.4 ENST00000510496.1 ENST00000380515.3 |
FAM169A |
family with sequence similarity 169, member A |
| chr6_+_64345698 | 0.94 |
ENST00000506783.1 ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3 |
PHD finger protein 3 |
| chr21_-_43735628 | 0.92 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr12_-_122985067 | 0.90 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chrY_+_15016013 | 0.89 |
ENST00000360160.4 ENST00000454054.1 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr5_+_156696362 | 0.88 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr9_-_5833027 | 0.88 |
ENST00000339450.5 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
| chr8_-_13372253 | 0.85 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr7_+_149571045 | 0.84 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr6_+_32709119 | 0.83 |
ENST00000374940.3 |
HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
| chr2_+_128177458 | 0.82 |
ENST00000409048.1 ENST00000422777.3 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr14_-_23426322 | 0.82 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr1_+_15943995 | 0.80 |
ENST00000480945.1 |
DDI2 |
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr4_+_128554081 | 0.80 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chrX_-_15511438 | 0.79 |
ENST00000380420.5 |
PIR |
pirin (iron-binding nuclear protein) |
| chr6_+_27791862 | 0.79 |
ENST00000355057.1 |
HIST1H4J |
histone cluster 1, H4j |
| chr5_-_42812143 | 0.78 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr11_+_121322832 | 0.78 |
ENST00000260197.7 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr6_-_155635583 | 0.78 |
ENST00000367166.4 |
TFB1M |
transcription factor B1, mitochondrial |
| chr6_-_26056695 | 0.78 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chrX_-_43741594 | 0.76 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr8_+_38585704 | 0.76 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_48189612 | 0.76 |
ENST00000453919.1 |
AGAP9 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr15_+_59903975 | 0.75 |
ENST00000560585.1 ENST00000396065.1 |
GCNT3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_-_75198940 | 0.75 |
ENST00000417775.1 |
CRYZ |
crystallin, zeta (quinone reductase) |
| chr19_-_43032532 | 0.75 |
ENST00000403461.1 ENST00000352591.5 ENST00000358394.3 ENST00000403444.3 ENST00000308072.4 ENST00000599389.1 ENST00000351134.3 ENST00000161559.6 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| chr16_+_10479906 | 0.74 |
ENST00000562527.1 ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2 |
activating transcription factor 7 interacting protein 2 |
| chr6_+_25652432 | 0.74 |
ENST00000377961.2 |
SCGN |
secretagogin, EF-hand calcium binding protein |
| chr10_+_57358750 | 0.73 |
ENST00000512524.2 |
MTRNR2L5 |
MT-RNR2-like 5 |
| chr19_+_52772832 | 0.73 |
ENST00000593703.1 ENST00000601711.1 ENST00000599581.1 |
ZNF766 |
zinc finger protein 766 |
| chr2_-_158345462 | 0.73 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr2_-_89399845 | 0.72 |
ENST00000479981.1 |
IGKV1-16 |
immunoglobulin kappa variable 1-16 |
| chr1_-_235814048 | 0.72 |
ENST00000450593.1 ENST00000366598.4 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
| chr4_+_90823130 | 0.72 |
ENST00000508372.1 |
MMRN1 |
multimerin 1 |
| chr18_-_67624160 | 0.71 |
ENST00000581982.1 ENST00000280200.4 |
CD226 |
CD226 molecule |
| chr11_+_7618413 | 0.70 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_64798095 | 0.69 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr17_+_67498538 | 0.68 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr1_+_70876926 | 0.68 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr10_-_115613828 | 0.68 |
ENST00000361384.2 |
DCLRE1A |
DNA cross-link repair 1A |
| chr8_-_19459993 | 0.68 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_+_97506033 | 0.68 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr11_+_128563652 | 0.68 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_105937068 | 0.67 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr18_+_32558208 | 0.67 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr18_+_76829441 | 0.67 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chr7_+_142498725 | 0.67 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr6_+_44215603 | 0.67 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr1_-_45140074 | 0.67 |
ENST00000420706.1 ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53 |
transmembrane protein 53 |
| chr9_-_36400920 | 0.66 |
ENST00000357058.3 ENST00000350199.4 |
RNF38 |
ring finger protein 38 |
| chr1_+_11333245 | 0.66 |
ENST00000376810.5 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
| chr8_-_17555164 | 0.65 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr5_-_94417339 | 0.65 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr20_+_30697298 | 0.64 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr19_+_42212501 | 0.63 |
ENST00000398599.4 |
CEACAM5 |
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr6_+_153552455 | 0.63 |
ENST00000392385.2 |
AL590867.1 |
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr1_-_111506562 | 0.62 |
ENST00000485275.2 ENST00000369763.4 |
LRIF1 |
ligand dependent nuclear receptor interacting factor 1 |
| chr21_-_15918618 | 0.61 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr8_-_13134045 | 0.61 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr1_+_201979645 | 0.60 |
ENST00000367284.5 ENST00000367283.3 |
ELF3 |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr6_+_31583761 | 0.60 |
ENST00000376049.4 |
AIF1 |
allograft inflammatory factor 1 |
| chr2_+_102953608 | 0.59 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr13_-_46756351 | 0.59 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chrX_+_51942963 | 0.59 |
ENST00000375625.3 |
RP11-363G10.2 |
RP11-363G10.2 |
| chr6_-_135271260 | 0.59 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr15_-_50558223 | 0.59 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr1_-_160492994 | 0.58 |
ENST00000368055.1 ENST00000368057.3 ENST00000368059.3 |
SLAMF6 |
SLAM family member 6 |
| chr21_+_44073860 | 0.58 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chr10_-_47239738 | 0.57 |
ENST00000413193.2 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr3_+_119501557 | 0.56 |
ENST00000337940.4 |
NR1I2 |
nuclear receptor subfamily 1, group I, member 2 |
| chr6_-_135271219 | 0.56 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr6_+_148663729 | 0.55 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chrX_+_38420623 | 0.55 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr10_+_105127704 | 0.55 |
ENST00000369839.3 ENST00000351396.4 |
TAF5 |
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
| chr11_-_62323702 | 0.54 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr6_+_150920999 | 0.54 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr5_+_121647386 | 0.53 |
ENST00000542191.1 ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr3_+_111697843 | 0.53 |
ENST00000534857.1 ENST00000273359.3 ENST00000494817.1 |
ABHD10 |
abhydrolase domain containing 10 |
| chr3_+_46618727 | 0.53 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr15_+_75498355 | 0.53 |
ENST00000567617.1 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr17_+_7155819 | 0.53 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr14_-_23426337 | 0.52 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr11_-_124670550 | 0.52 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr1_+_70820451 | 0.52 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr4_-_105416039 | 0.52 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr6_+_32605195 | 0.52 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr21_-_43735446 | 0.52 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr1_+_11994715 | 0.51 |
ENST00000449038.1 ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr21_+_44073916 | 0.51 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr17_-_46507567 | 0.51 |
ENST00000584924.1 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr20_-_47804894 | 0.51 |
ENST00000371828.3 ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
| chr6_+_32605134 | 0.51 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chrX_-_38186811 | 0.51 |
ENST00000318842.7 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr1_+_160709055 | 0.50 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chrX_-_107975917 | 0.50 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chrX_-_16730688 | 0.50 |
ENST00000359276.4 |
CTPS2 |
CTP synthase 2 |
| chr15_-_65426174 | 0.50 |
ENST00000204549.4 |
PDCD7 |
programmed cell death 7 |
| chr1_-_212965104 | 0.49 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chr1_+_160709029 | 0.49 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chr20_+_45338126 | 0.49 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr22_-_29107919 | 0.48 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr5_+_95066823 | 0.48 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr6_+_26204825 | 0.48 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chrX_+_38420783 | 0.48 |
ENST00000422612.2 ENST00000286824.6 ENST00000545599.1 |
TSPAN7 |
tetraspanin 7 |
| chr5_-_169725231 | 0.48 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr2_-_191115229 | 0.47 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_+_150020329 | 0.47 |
ENST00000323078.7 |
LRRC61 |
leucine rich repeat containing 61 |
| chr6_+_106546808 | 0.47 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr11_-_128894053 | 0.47 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr9_+_130159409 | 0.47 |
ENST00000373371.3 |
SLC2A8 |
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr10_-_115614127 | 0.47 |
ENST00000369305.1 |
DCLRE1A |
DNA cross-link repair 1A |
| chr1_-_150738261 | 0.47 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr16_+_70258261 | 0.46 |
ENST00000594734.1 |
FKSG63 |
FKSG63 |
| chr15_+_58724184 | 0.45 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr7_+_150020363 | 0.45 |
ENST00000359623.4 ENST00000493307.1 |
LRRC61 |
leucine rich repeat containing 61 |
| chr3_+_98482175 | 0.44 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr11_+_95523823 | 0.44 |
ENST00000538658.1 |
CEP57 |
centrosomal protein 57kDa |
| chr2_+_108905325 | 0.44 |
ENST00000438339.1 ENST00000409880.1 ENST00000437390.2 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_+_139528952 | 0.43 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr7_-_20256965 | 0.43 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr2_+_89890533 | 0.43 |
ENST00000429992.2 |
IGKV2D-40 |
immunoglobulin kappa variable 2D-40 |
| chr15_-_47426320 | 0.43 |
ENST00000557832.1 |
FKSG62 |
FKSG62 |
| chr4_-_101439148 | 0.43 |
ENST00000511970.1 ENST00000502569.1 ENST00000305864.3 |
EMCN |
endomucin |
| chrX_-_38186775 | 0.42 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr19_+_54466179 | 0.42 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr14_+_31091511 | 0.42 |
ENST00000544052.2 ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1 |
sec1 family domain containing 1 |
| chr17_-_29641104 | 0.42 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr1_-_11918988 | 0.42 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr9_-_123691439 | 0.42 |
ENST00000540010.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chrX_-_16730984 | 0.42 |
ENST00000380241.3 |
CTPS2 |
CTP synthase 2 |
| chr2_+_73612858 | 0.42 |
ENST00000409009.1 ENST00000264448.6 ENST00000377715.1 |
ALMS1 |
Alstrom syndrome 1 |
| chr7_+_97361218 | 0.42 |
ENST00000319273.5 |
TAC1 |
tachykinin, precursor 1 |
| chr17_-_46507537 | 0.41 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr1_-_205601064 | 0.41 |
ENST00000357992.4 ENST00000289703.4 |
ELK4 |
ELK4, ETS-domain protein (SRF accessory protein 1) |
| chr19_+_40194946 | 0.41 |
ENST00000392052.3 |
LGALS14 |
lectin, galactoside-binding, soluble, 14 |
| chr1_+_12976450 | 0.41 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr6_-_52149475 | 0.41 |
ENST00000419835.2 ENST00000229854.7 ENST00000596288.1 |
MCM3 |
minichromosome maintenance complex component 3 |
| chr5_+_175288631 | 0.41 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr2_-_111435610 | 0.41 |
ENST00000447014.1 ENST00000420328.1 ENST00000535254.1 ENST00000409311.1 ENST00000302759.6 |
BUB1 |
BUB1 mitotic checkpoint serine/threonine kinase |
| chr16_-_21431078 | 0.41 |
ENST00000458643.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr16_+_20775024 | 0.41 |
ENST00000289416.5 |
ACSM3 |
acyl-CoA synthetase medium-chain family member 3 |
| chr3_+_173116225 | 0.41 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr20_+_2854066 | 0.40 |
ENST00000455631.1 ENST00000216877.6 ENST00000399903.2 ENST00000358719.4 ENST00000431048.1 ENST00000425918.2 ENST00000430705.1 ENST00000318266.5 |
PTPRA |
protein tyrosine phosphatase, receptor type, A |
| chr18_+_10526008 | 0.40 |
ENST00000542979.1 ENST00000322897.6 |
NAPG |
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr2_-_89417335 | 0.40 |
ENST00000490686.1 |
IGKV1-17 |
immunoglobulin kappa variable 1-17 |
| chr17_-_29641084 | 0.40 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr15_-_55563072 | 0.40 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr2_+_234826016 | 0.40 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr19_+_40195101 | 0.40 |
ENST00000360675.3 ENST00000601802.1 |
LGALS14 |
lectin, galactoside-binding, soluble, 14 |
| chr5_+_154393260 | 0.39 |
ENST00000435029.4 |
KIF4B |
kinesin family member 4B |
| chr3_+_135969148 | 0.39 |
ENST00000251654.4 ENST00000490504.1 ENST00000483687.1 ENST00000468777.1 ENST00000462637.1 ENST00000466072.1 ENST00000482086.1 ENST00000471595.1 ENST00000469217.1 ENST00000465423.1 ENST00000478469.1 |
PCCB |
propionyl CoA carboxylase, beta polypeptide |
| chr2_-_89292422 | 0.39 |
ENST00000495489.1 |
IGKV1-8 |
immunoglobulin kappa variable 1-8 |
| chr11_+_43380459 | 0.39 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr11_+_102188224 | 0.39 |
ENST00000263464.3 |
BIRC3 |
baculoviral IAP repeat containing 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0034255 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.9 | 2.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.5 | 1.6 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.4 | 4.9 | GO:0051918 | positive regulation of lipoprotein lipase activity(GO:0051006) negative regulation of fibrinolysis(GO:0051918) |
| 0.4 | 2.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 0.8 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.3 | 0.8 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.7 | GO:0043318 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) insulin catabolic process(GO:1901143) |
| 0.2 | 0.7 | GO:0060369 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 2.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 2.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 4.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.2 | 4.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.6 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.2 | 2.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 1.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 1.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.8 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.2 | 0.7 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 1.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.7 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 1.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) NMDA glutamate receptor clustering(GO:0097114) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.7 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.1 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.1 | 3.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.8 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.7 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 0.6 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.3 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.2 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.2 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 1.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:2000566 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.5 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.2 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 1.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.8 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.4 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 1.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.7 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.1 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.8 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.1 | 0.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) regulation of catecholamine secretion(GO:0050433) |
| 0.1 | 0.3 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.3 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.4 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 6.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.5 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.9 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 1.0 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.7 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.4 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.4 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.5 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.5 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 1.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.0 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.4 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0007618 | mating(GO:0007618) |
| 0.0 | 0.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.0 | 3.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.5 | 1.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 2.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 10.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 0.7 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 2.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 1.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 2.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.2 | 1.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.7 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 2.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) mediator complex binding(GO:0036033) |
| 0.1 | 0.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 2.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 1.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 3.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 3.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0044388 | ubiquitin activating enzyme activity(GO:0004839) small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.6 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.1 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 4.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.3 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 2.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.2 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 1.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 7.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |