Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
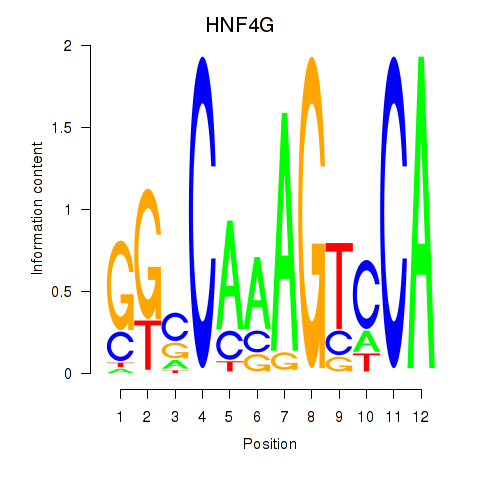
Results for HNF4G
Z-value: 1.68
Transcription factors associated with HNF4G
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4G
|
ENSG00000164749.7 | HNF4G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4G | hg19_v2_chr8_+_76452097_76452126 | 0.25 | 3.4e-01 | Click! |
Activity profile of HNF4G motif
Sorted Z-values of HNF4G motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4G
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_6720686 | 4.65 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr17_-_7082668 | 4.48 |
ENST00000573083.1 ENST00000574388.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr17_-_7082861 | 3.73 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr1_-_169555779 | 3.52 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr17_+_27369918 | 3.50 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chr11_-_2158507 | 3.33 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr13_+_113760098 | 3.12 |
ENST00000346342.3 ENST00000541084.1 ENST00000375581.3 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr14_-_94854926 | 3.12 |
ENST00000402629.1 ENST00000556091.1 ENST00000554720.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr8_-_27468842 | 3.01 |
ENST00000523500.1 |
CLU |
clusterin |
| chr8_-_27469196 | 2.85 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr19_+_18496957 | 2.84 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr2_-_238499303 | 2.83 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr16_-_56701933 | 2.83 |
ENST00000568675.1 ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G |
metallothionein 1G |
| chrX_+_51927919 | 2.81 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr3_-_123339418 | 2.81 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr1_-_151965048 | 2.75 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr3_-_123339343 | 2.69 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr9_+_124030338 | 2.61 |
ENST00000449773.1 ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN |
gelsolin |
| chr13_+_113777105 | 2.54 |
ENST00000409306.1 ENST00000375551.3 ENST00000375559.3 |
F10 |
coagulation factor X |
| chr2_-_161350305 | 2.51 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr16_+_56659687 | 2.47 |
ENST00000568293.1 ENST00000330439.6 |
MT1E |
metallothionein 1E |
| chr19_+_45449301 | 2.44 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr19_+_45449228 | 2.44 |
ENST00000252490.4 |
APOC2 |
apolipoprotein C-II |
| chr9_+_133320339 | 2.43 |
ENST00000372394.1 ENST00000372393.3 ENST00000422569.1 |
ASS1 |
argininosuccinate synthase 1 |
| chr19_+_45449266 | 2.39 |
ENST00000592257.1 |
APOC2 |
apolipoprotein C-II |
| chr7_+_45927956 | 2.34 |
ENST00000275525.3 ENST00000457280.1 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
| chr8_-_27468945 | 2.31 |
ENST00000405140.3 |
CLU |
clusterin |
| chr12_+_56477093 | 2.26 |
ENST00000549672.1 ENST00000415288.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chrX_-_130423386 | 2.25 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr7_+_45928079 | 2.22 |
ENST00000468955.1 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
| chr13_+_27131887 | 2.19 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chr1_+_207262578 | 2.17 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr11_+_116700614 | 2.12 |
ENST00000375345.1 |
APOC3 |
apolipoprotein C-III |
| chrX_-_99891796 | 2.10 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr2_-_20212422 | 2.02 |
ENST00000421259.2 ENST00000407540.3 |
MATN3 |
matrilin 3 |
| chr18_+_7754957 | 1.99 |
ENST00000400053.4 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr2_+_201170596 | 1.99 |
ENST00000439084.1 ENST00000409718.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr3_-_185542761 | 1.98 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_-_52488048 | 1.97 |
ENST00000232975.3 |
TNNC1 |
troponin C type 1 (slow) |
| chr3_-_185542817 | 1.95 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr16_+_3115378 | 1.95 |
ENST00000529550.1 ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32 |
interleukin 32 |
| chr2_+_219283815 | 1.89 |
ENST00000248444.5 ENST00000454069.1 ENST00000392114.2 |
VIL1 |
villin 1 |
| chr11_+_116700600 | 1.87 |
ENST00000227667.3 |
APOC3 |
apolipoprotein C-III |
| chrX_-_43741594 | 1.86 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr6_+_31620191 | 1.85 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr20_+_48807351 | 1.83 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr4_+_6271558 | 1.78 |
ENST00000503569.1 ENST00000226760.1 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
| chr2_+_201170770 | 1.76 |
ENST00000409988.3 ENST00000409385.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr14_-_53331239 | 1.68 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr11_+_12399071 | 1.64 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr14_+_52781079 | 1.64 |
ENST00000245457.5 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr1_+_145727681 | 1.64 |
ENST00000417171.1 ENST00000451928.2 |
PDZK1 |
PDZ domain containing 1 |
| chr2_+_85811525 | 1.63 |
ENST00000306384.4 |
VAMP5 |
vesicle-associated membrane protein 5 |
| chr1_+_78511586 | 1.61 |
ENST00000370759.3 |
GIPC2 |
GIPC PDZ domain containing family, member 2 |
| chr1_+_155099927 | 1.60 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr16_+_56642041 | 1.60 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr18_+_3412005 | 1.59 |
ENST00000401449.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr7_-_87104963 | 1.57 |
ENST00000359206.3 ENST00000358400.3 ENST00000265723.4 |
ABCB4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_236686875 | 1.54 |
ENST00000366584.4 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr16_+_3115611 | 1.54 |
ENST00000530890.1 ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32 |
interleukin 32 |
| chr10_-_69597915 | 1.53 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr3_+_138068051 | 1.52 |
ENST00000474559.1 |
MRAS |
muscle RAS oncogene homolog |
| chr8_+_22436635 | 1.51 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr16_+_3115298 | 1.50 |
ENST00000325568.5 ENST00000534507.1 |
IL32 |
interleukin 32 |
| chr2_-_238499725 | 1.45 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr6_+_30130969 | 1.43 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr1_+_153600869 | 1.41 |
ENST00000292169.1 ENST00000368696.3 ENST00000436839.1 |
S100A1 |
S100 calcium binding protein A1 |
| chr20_+_34680620 | 1.41 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr20_+_61584026 | 1.40 |
ENST00000370351.4 ENST00000370349.3 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr2_-_238499131 | 1.39 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr10_-_69597810 | 1.31 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_+_207262540 | 1.29 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr8_-_11710979 | 1.28 |
ENST00000415599.2 |
CTSB |
cathepsin B |
| chr2_+_241807870 | 1.27 |
ENST00000307503.3 |
AGXT |
alanine-glyoxylate aminotransferase |
| chr16_+_55512742 | 1.27 |
ENST00000568715.1 ENST00000219070.4 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr16_-_75467318 | 1.26 |
ENST00000283882.3 |
CFDP1 |
craniofacial development protein 1 |
| chr6_+_131894284 | 1.25 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chrX_+_46937745 | 1.25 |
ENST00000397180.1 ENST00000457380.1 ENST00000352078.4 |
RGN |
regucalcin |
| chr1_+_180165672 | 1.24 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr20_+_19867150 | 1.24 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr16_-_87970122 | 1.21 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr7_-_94285402 | 1.21 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr8_+_11561660 | 1.20 |
ENST00000526716.1 ENST00000335135.4 ENST00000528027.1 |
GATA4 |
GATA binding protein 4 |
| chr12_+_6309963 | 1.20 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr1_+_207262170 | 1.19 |
ENST00000367078.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr2_+_201170703 | 1.17 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr2_-_238499337 | 1.17 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr17_+_80186908 | 1.16 |
ENST00000582743.1 ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_3069037 | 1.16 |
ENST00000397122.2 |
TEAD4 |
TEA domain family member 4 |
| chr16_-_89768097 | 1.15 |
ENST00000289805.5 ENST00000335360.7 |
SPATA2L |
spermatogenesis associated 2-like |
| chr3_+_138067521 | 1.14 |
ENST00000494949.1 |
MRAS |
muscle RAS oncogene homolog |
| chr1_+_17906970 | 1.12 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_-_94285472 | 1.12 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr17_+_48624450 | 1.11 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr5_+_34757309 | 1.10 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr14_-_24911971 | 1.10 |
ENST00000555365.1 ENST00000399395.3 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr11_-_66725837 | 1.09 |
ENST00000393958.2 ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC |
pyruvate carboxylase |
| chr2_-_47168906 | 1.06 |
ENST00000444761.2 ENST00000409147.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr12_-_81331460 | 1.05 |
ENST00000549417.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chr16_+_3115323 | 1.05 |
ENST00000531965.1 ENST00000396887.3 ENST00000529699.1 ENST00000526464.2 ENST00000440815.3 |
IL32 |
interleukin 32 |
| chr11_+_32112431 | 1.04 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr7_-_94285511 | 1.03 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr11_+_1860200 | 1.02 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr14_+_101302041 | 1.02 |
ENST00000522618.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr12_+_121416489 | 1.02 |
ENST00000541395.1 ENST00000544413.1 |
HNF1A |
HNF1 homeobox A |
| chr6_+_146864829 | 0.99 |
ENST00000367495.3 |
RAB32 |
RAB32, member RAS oncogene family |
| chr1_-_153508460 | 0.98 |
ENST00000462776.2 |
S100A6 |
S100 calcium binding protein A6 |
| chr16_-_66764119 | 0.97 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr2_+_241544834 | 0.95 |
ENST00000319838.5 ENST00000403859.1 ENST00000438013.2 |
GPR35 |
G protein-coupled receptor 35 |
| chr12_+_10366016 | 0.95 |
ENST00000546017.1 ENST00000535576.1 ENST00000539170.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr22_+_38093005 | 0.94 |
ENST00000406386.3 |
TRIOBP |
TRIO and F-actin binding protein |
| chr4_-_16077741 | 0.94 |
ENST00000447510.2 ENST00000540805.1 ENST00000539194.1 |
PROM1 |
prominin 1 |
| chr16_+_57406368 | 0.94 |
ENST00000006053.6 ENST00000563383.1 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
| chr20_+_43211149 | 0.93 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr9_-_79307096 | 0.93 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr1_-_153518270 | 0.92 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr3_-_46000064 | 0.92 |
ENST00000433878.1 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr11_-_111781554 | 0.92 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr11_+_67070919 | 0.92 |
ENST00000308127.4 ENST00000308298.7 |
SSH3 |
slingshot protein phosphatase 3 |
| chr2_+_191208196 | 0.92 |
ENST00000392329.2 ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1 |
inositol polyphosphate-1-phosphatase |
| chr11_+_67071050 | 0.92 |
ENST00000376757.5 |
SSH3 |
slingshot protein phosphatase 3 |
| chr14_+_55595762 | 0.91 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr11_-_111781610 | 0.90 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr19_-_34012674 | 0.90 |
ENST00000436370.3 ENST00000397032.4 ENST00000244137.7 |
PEPD |
peptidase D |
| chr2_+_217524323 | 0.88 |
ENST00000456764.1 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr11_-_111783595 | 0.87 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chrX_-_15288154 | 0.86 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chr11_+_1860832 | 0.86 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr8_-_38326119 | 0.84 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr2_-_20251744 | 0.83 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr8_+_27491572 | 0.81 |
ENST00000301904.3 |
SCARA3 |
scavenger receptor class A, member 3 |
| chr14_+_62162258 | 0.81 |
ENST00000337138.4 ENST00000394997.1 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr1_-_201390846 | 0.80 |
ENST00000367312.1 ENST00000555340.2 ENST00000361379.4 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr12_+_13197218 | 0.80 |
ENST00000197268.8 |
KIAA1467 |
KIAA1467 |
| chr3_+_122785895 | 0.79 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chrX_+_54834004 | 0.79 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr10_+_89419370 | 0.79 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr18_+_19749386 | 0.78 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr7_-_27170352 | 0.77 |
ENST00000428284.2 ENST00000360046.5 |
HOXA4 |
homeobox A4 |
| chr11_-_111783919 | 0.76 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_145642267 | 0.76 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr5_+_148521381 | 0.76 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr22_-_43045574 | 0.76 |
ENST00000352397.5 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr1_+_246887349 | 0.75 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr9_-_130639997 | 0.75 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr2_+_54785485 | 0.74 |
ENST00000333896.5 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr18_-_52626622 | 0.74 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr8_-_38326139 | 0.73 |
ENST00000335922.5 ENST00000532791.1 ENST00000397091.5 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr8_-_145159083 | 0.72 |
ENST00000398712.2 |
SHARPIN |
SHANK-associated RH domain interactor |
| chr2_-_47168850 | 0.71 |
ENST00000409207.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr11_-_63536113 | 0.70 |
ENST00000433688.1 ENST00000546282.2 |
C11orf95 RP11-466C23.4 |
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chrX_-_48693955 | 0.67 |
ENST00000218230.5 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr13_+_113812956 | 0.67 |
ENST00000375547.2 ENST00000342783.4 |
PROZ |
protein Z, vitamin K-dependent plasma glycoprotein |
| chr3_+_38495333 | 0.67 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr12_-_56882136 | 0.66 |
ENST00000311966.4 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
| chr12_+_112204691 | 0.66 |
ENST00000416293.3 ENST00000261733.2 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr16_+_3493611 | 0.66 |
ENST00000407558.4 ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr17_+_7482785 | 0.66 |
ENST00000250092.6 ENST00000380498.6 ENST00000584502.1 |
CD68 |
CD68 molecule |
| chr8_+_21911054 | 0.65 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr15_+_99645277 | 0.65 |
ENST00000336292.6 ENST00000328642.7 |
SYNM |
synemin, intermediate filament protein |
| chr11_+_1860682 | 0.65 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr17_-_38256973 | 0.65 |
ENST00000246672.3 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
| chr14_-_24911868 | 0.65 |
ENST00000554698.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr11_-_111781454 | 0.65 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr3_+_52812523 | 0.65 |
ENST00000540715.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr14_+_55595960 | 0.65 |
ENST00000554715.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr7_-_121784285 | 0.64 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr17_+_68165657 | 0.64 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr20_+_62694461 | 0.63 |
ENST00000343484.5 ENST00000395053.3 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr19_-_4400415 | 0.63 |
ENST00000598564.1 ENST00000417295.2 ENST00000269886.3 |
SH3GL1 |
SH3-domain GRB2-like 1 |
| chr2_-_242447983 | 0.62 |
ENST00000426941.1 ENST00000405585.1 ENST00000420551.1 ENST00000535007.1 ENST00000429279.1 ENST00000442307.1 ENST00000403346.3 ENST00000316586.4 |
STK25 |
serine/threonine kinase 25 |
| chr11_-_111784005 | 0.61 |
ENST00000527899.1 |
CRYAB |
crystallin, alpha B |
| chr22_-_37415475 | 0.61 |
ENST00000403892.3 ENST00000249042.3 ENST00000438203.1 |
TST |
thiosulfate sulfurtransferase (rhodanese) |
| chr8_-_145641864 | 0.61 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr11_+_111783450 | 0.61 |
ENST00000537382.1 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr17_-_17740287 | 0.60 |
ENST00000355815.4 ENST00000261646.5 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
| chr1_-_201391149 | 0.60 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr22_+_37415700 | 0.60 |
ENST00000397129.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr1_+_21766588 | 0.59 |
ENST00000454000.2 ENST00000318220.6 ENST00000318249.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr22_+_37415776 | 0.59 |
ENST00000341116.3 ENST00000429360.2 ENST00000404393.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr3_-_47950745 | 0.58 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr15_-_72668805 | 0.58 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr2_-_70475730 | 0.58 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr14_-_24664540 | 0.55 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr3_+_138067314 | 0.55 |
ENST00000423968.2 |
MRAS |
muscle RAS oncogene homolog |
| chr5_+_179125907 | 0.54 |
ENST00000247461.4 ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX |
calnexin |
| chrX_+_2746850 | 0.53 |
ENST00000381163.3 ENST00000338623.5 ENST00000542787.1 |
GYG2 |
glycogenin 2 |
| chr3_+_37284824 | 0.53 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr2_-_70475701 | 0.53 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr12_-_90049878 | 0.52 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_-_220110111 | 0.52 |
ENST00000428427.1 ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L |
galactosidase, beta 1-like |
| chr11_+_842808 | 0.52 |
ENST00000397397.2 ENST00000397411.2 ENST00000397396.1 |
TSPAN4 |
tetraspanin 4 |
| chr3_+_138067666 | 0.52 |
ENST00000475711.1 ENST00000464896.1 |
MRAS |
muscle RAS oncogene homolog |
| chr17_-_73975444 | 0.51 |
ENST00000293217.5 ENST00000537812.1 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
| chr5_-_131330272 | 0.50 |
ENST00000379240.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr22_+_45072958 | 0.50 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr19_-_14629224 | 0.50 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_-_120612240 | 0.49 |
ENST00000256646.2 |
NOTCH2 |
notch 2 |
| chr22_+_37415728 | 0.49 |
ENST00000404802.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr6_+_108977520 | 0.48 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 1.5 | 4.6 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 1.4 | 4.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 1.4 | 6.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.4 | 8.2 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.3 | 6.6 | GO:0032571 | response to vitamin K(GO:0032571) |
| 1.3 | 4.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.7 | 5.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 0.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 1.7 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.5 | 2.7 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.5 | 1.6 | GO:0097327 | response to thyroxine(GO:0097068) response to antineoplastic agent(GO:0097327) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) response to L-phenylalanine derivative(GO:1904386) |
| 0.5 | 1.6 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.5 | 2.6 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.5 | 1.6 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.5 | 4.7 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.5 | 2.0 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.5 | 1.5 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.4 | 3.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.4 | 1.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.4 | 1.6 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.4 | 1.2 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.4 | 1.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 1.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 3.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 1.7 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 1.6 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 3.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.3 | 0.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.3 | 2.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 1.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.3 | 1.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 0.8 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 1.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 4.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 0.7 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.2 | 0.6 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 | 1.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 1.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.2 | 0.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.4 | GO:0032058 | positive regulation of translation in response to stress(GO:0032056) positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 0.6 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.2 | 0.9 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 0.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 1.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 10.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.7 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 2.7 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.5 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.2 | 0.9 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 2.1 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 | 2.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 4.4 | GO:0036499 | PERK-mediated unfolded protein response(GO:0036499) |
| 0.1 | 1.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 1.8 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.1 | GO:0090346 | cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 1.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 2.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 2.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.4 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 2.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.6 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 0.8 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.2 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 0.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 1.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.4 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.5 | GO:0060510 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) Type II pneumocyte differentiation(GO:0060510) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.8 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.2 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 2.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.7 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.3 | GO:0097067 | decidualization(GO:0046697) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.2 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.1 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.6 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.1 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 0.9 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 3.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.7 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.9 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 2.7 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 2.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 1.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 3.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:1902956 | negative regulation of cellular respiration(GO:1901856) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0042107 | cytokine metabolic process(GO:0042107) |
| 0.0 | 0.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.1 | 5.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 8.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.0 | 4.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.6 | 3.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 2.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.5 | 5.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 1.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.4 | 8.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 1.5 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.4 | 1.8 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 4.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 0.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 3.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 2.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 0.8 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.0 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 2.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 1.6 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 2.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.5 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 1.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 3.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.5 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 2.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.1 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.4 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 2.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 3.3 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 4.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 3.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 8.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 6.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 2.1 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 6.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 9.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 7.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 6.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 6.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 10.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 9.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 3.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 12.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 1.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.4 | 11.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.8 | 8.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 3.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 4.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 3.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.2 | 1.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 2.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 11.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.3 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 7.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 5.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 7.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 19.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 2.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.9 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.4 | GO:0030056 | apicolateral plasma membrane(GO:0016327) hemidesmosome(GO:0030056) |
| 0.0 | 3.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.9 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |