Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
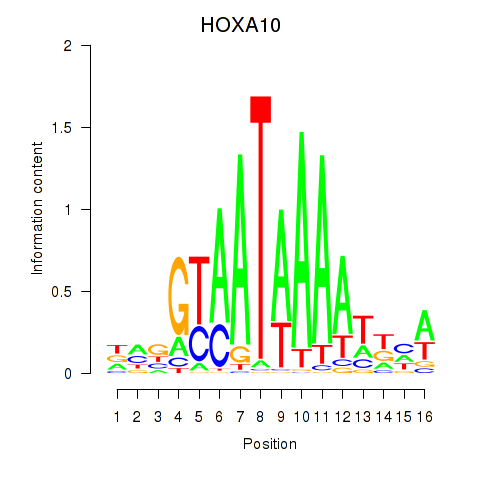
Results for HOXA10_HOXB9
Z-value: 1.24
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA10
|
ENSG00000253293.3 | HOXA10 |
|
HOXB9
|
ENSG00000170689.8 | HOXB9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27219849_27219880 | -0.55 | 2.7e-02 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | 0.45 | 7.9e-02 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_55541227 | 4.53 |
ENST00000566877.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr6_+_12958137 | 3.50 |
ENST00000457702.2 ENST00000379345.2 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr15_-_58571445 | 2.36 |
ENST00000558231.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr4_-_71532339 | 2.27 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr9_+_2029019 | 2.21 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_81083890 | 2.06 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chr4_+_68424434 | 1.75 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr6_+_6588316 | 1.53 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr11_+_22688150 | 1.45 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr12_+_54892550 | 1.44 |
ENST00000545638.2 |
NCKAP1L |
NCK-associated protein 1-like |
| chr1_+_12524965 | 1.39 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_-_58196688 | 1.38 |
ENST00000486455.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr12_+_25205568 | 1.31 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr1_+_241695424 | 1.19 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr12_-_8765446 | 1.16 |
ENST00000537228.1 ENST00000229335.6 |
AICDA |
activation-induced cytidine deaminase |
| chr10_-_73848531 | 1.15 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr3_+_157828152 | 1.14 |
ENST00000476899.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chr12_+_25205446 | 1.04 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr6_+_26156551 | 1.04 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chrX_-_73072534 | 1.03 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr12_-_57146095 | 0.99 |
ENST00000550770.1 ENST00000338193.6 |
PRIM1 |
primase, DNA, polypeptide 1 (49kDa) |
| chr1_+_74701062 | 0.97 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr9_+_134065506 | 0.95 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr10_-_115614127 | 0.95 |
ENST00000369305.1 |
DCLRE1A |
DNA cross-link repair 1A |
| chr4_-_76957214 | 0.95 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr22_-_23922410 | 0.94 |
ENST00000249053.3 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr13_-_47012325 | 0.92 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chrX_-_135962876 | 0.88 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr12_+_25205666 | 0.88 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr8_+_77593448 | 0.86 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr6_+_44215603 | 0.85 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr3_+_169629354 | 0.83 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
| chr6_-_166582107 | 0.79 |
ENST00000296946.2 ENST00000461348.2 ENST00000366871.3 |
T |
T, brachyury homolog (mouse) |
| chr5_-_88119580 | 0.78 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr6_+_42584847 | 0.78 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr8_-_81083731 | 0.77 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr5_+_67586465 | 0.77 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_-_39339777 | 0.73 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr11_-_3400442 | 0.72 |
ENST00000429541.2 ENST00000532539.1 |
ZNF195 |
zinc finger protein 195 |
| chr12_+_1800179 | 0.72 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr4_-_100356551 | 0.72 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_+_43121698 | 0.71 |
ENST00000505606.2 ENST00000509634.1 ENST00000509341.1 |
ZNF131 |
zinc finger protein 131 |
| chr1_+_241695670 | 0.70 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr5_+_34915444 | 0.69 |
ENST00000336767.5 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr11_-_3400330 | 0.69 |
ENST00000427810.2 ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195 |
zinc finger protein 195 |
| chr11_+_63606373 | 0.67 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chrX_-_100604184 | 0.65 |
ENST00000372902.3 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr17_-_39646116 | 0.61 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr5_-_58295712 | 0.60 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr11_-_118972575 | 0.60 |
ENST00000432443.2 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr2_-_51259229 | 0.59 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chrX_+_65384182 | 0.59 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chrX_-_122756660 | 0.58 |
ENST00000441692.1 |
THOC2 |
THO complex 2 |
| chrX_-_133792480 | 0.58 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr1_+_153003671 | 0.57 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr10_+_102106829 | 0.57 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr10_-_73848086 | 0.57 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr3_+_133118839 | 0.56 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr9_+_42671887 | 0.55 |
ENST00000456520.1 ENST00000377391.3 |
CBWD7 |
COBW domain containing 7 |
| chr1_+_23345943 | 0.55 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr2_-_176046391 | 0.55 |
ENST00000392541.3 ENST00000409194.1 |
ATP5G3 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr12_-_54071181 | 0.55 |
ENST00000338662.5 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr1_+_23345930 | 0.54 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr8_+_95565947 | 0.53 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr7_-_140482926 | 0.52 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr15_-_22448819 | 0.51 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr2_-_51259292 | 0.51 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chrX_-_110655306 | 0.50 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr2_+_234668894 | 0.49 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_27440229 | 0.49 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_102576537 | 0.48 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr9_-_179018 | 0.47 |
ENST00000431099.2 ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1 |
COBW domain containing 1 |
| chr2_+_90198535 | 0.47 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chrX_-_15619076 | 0.46 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr8_+_77593474 | 0.46 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr10_+_94451574 | 0.46 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr6_-_109804412 | 0.46 |
ENST00000230122.3 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
| chr7_+_57509877 | 0.46 |
ENST00000420713.1 |
ZNF716 |
zinc finger protein 716 |
| chrX_+_65384052 | 0.45 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr20_+_42984330 | 0.45 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr11_+_10471836 | 0.45 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr8_-_95220775 | 0.44 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr5_+_140261703 | 0.44 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr1_+_67773044 | 0.43 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr2_-_40657397 | 0.41 |
ENST00000408028.2 ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr12_-_71148413 | 0.40 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr20_+_10015678 | 0.39 |
ENST00000378392.1 ENST00000378380.3 |
ANKEF1 |
ankyrin repeat and EF-hand domain containing 1 |
| chr12_-_71148357 | 0.39 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr19_+_1440838 | 0.39 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr1_-_186430222 | 0.38 |
ENST00000391997.2 |
PDC |
phosducin |
| chr21_+_35736302 | 0.38 |
ENST00000290310.3 |
KCNE2 |
potassium voltage-gated channel, Isk-related family, member 2 |
| chr1_+_117963209 | 0.38 |
ENST00000449370.2 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
| chr6_+_46761118 | 0.36 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr1_+_52682052 | 0.35 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr3_+_189507432 | 0.35 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr1_+_2985726 | 0.35 |
ENST00000511072.1 ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16 |
PR domain containing 16 |
| chr15_+_66797455 | 0.34 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr9_-_95087838 | 0.34 |
ENST00000442668.2 ENST00000421075.2 ENST00000536624.1 |
NOL8 |
nucleolar protein 8 |
| chr3_+_15045419 | 0.34 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr7_-_7782204 | 0.34 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chr6_+_32812568 | 0.34 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr18_+_10526008 | 0.33 |
ENST00000542979.1 ENST00000322897.6 |
NAPG |
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr15_+_66797627 | 0.33 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr20_+_4702548 | 0.32 |
ENST00000305817.2 |
PRND |
prion protein 2 (dublet) |
| chr4_+_71263599 | 0.32 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr17_-_33469299 | 0.32 |
ENST00000586869.1 ENST00000360831.5 ENST00000442241.4 |
NLE1 |
notchless homolog 1 (Drosophila) |
| chr5_-_130500922 | 0.32 |
ENST00000513012.1 ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1 |
histidine triad nucleotide binding protein 1 |
| chr9_-_104145795 | 0.32 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr7_-_80141328 | 0.30 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr11_+_92085262 | 0.30 |
ENST00000298047.6 ENST00000409404.2 ENST00000541502.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr17_+_7211656 | 0.30 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr8_-_8318847 | 0.29 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr9_-_69262509 | 0.29 |
ENST00000377449.1 ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6 |
COBW domain containing 6 |
| chr12_-_18243075 | 0.29 |
ENST00000536890.1 |
RERGL |
RERG/RAS-like |
| chr12_-_18243119 | 0.29 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr17_-_46806540 | 0.29 |
ENST00000290295.7 |
HOXB13 |
homeobox B13 |
| chr7_-_143105941 | 0.28 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr14_-_106453155 | 0.27 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr8_+_76452097 | 0.27 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr19_+_4402659 | 0.27 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr13_-_103719196 | 0.27 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr22_-_23922448 | 0.26 |
ENST00000438703.1 ENST00000330377.2 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr7_-_25268104 | 0.26 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr10_-_103599591 | 0.26 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr5_-_176433693 | 0.26 |
ENST00000507513.1 ENST00000511320.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr20_-_7921090 | 0.26 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr14_-_60097297 | 0.26 |
ENST00000395090.1 |
RTN1 |
reticulon 1 |
| chr12_-_118628350 | 0.26 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr14_-_51027838 | 0.26 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_68962805 | 0.25 |
ENST00000370966.5 |
DEPDC1 |
DEP domain containing 1 |
| chr1_-_152386732 | 0.25 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr15_+_93749295 | 0.25 |
ENST00000599897.1 |
AC112693.2 |
AC112693.2 |
| chr6_-_26250835 | 0.25 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr20_+_30697298 | 0.25 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr15_+_67835517 | 0.25 |
ENST00000395476.2 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr12_-_24102576 | 0.25 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr2_+_208423840 | 0.24 |
ENST00000539789.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr6_-_137539651 | 0.24 |
ENST00000543628.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr17_-_39553844 | 0.24 |
ENST00000251645.2 |
KRT31 |
keratin 31 |
| chr11_+_120973375 | 0.24 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr11_-_104972158 | 0.23 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chrX_-_80457385 | 0.23 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr19_+_19626531 | 0.23 |
ENST00000507754.4 |
NDUFA13 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr3_-_196911002 | 0.23 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr19_+_21265028 | 0.23 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
| chr18_+_42260861 | 0.23 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr9_-_21202204 | 0.22 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr19_-_29704448 | 0.22 |
ENST00000304863.4 |
UQCRFS1 |
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr18_+_3252265 | 0.21 |
ENST00000580887.1 ENST00000536605.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr4_+_113568207 | 0.21 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr22_-_22337146 | 0.21 |
ENST00000398793.2 |
TOP3B |
topoisomerase (DNA) III beta |
| chr9_+_70856397 | 0.21 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr2_+_208423891 | 0.20 |
ENST00000448277.1 ENST00000457101.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr15_+_52155001 | 0.20 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr1_+_154401791 | 0.20 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chrX_+_70503433 | 0.20 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr16_+_72090053 | 0.20 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr20_-_34287103 | 0.20 |
ENST00000374085.1 ENST00000419569.1 |
NFS1 |
NFS1 cysteine desulfurase |
| chr4_-_170897045 | 0.20 |
ENST00000508313.1 |
RP11-205M3.3 |
RP11-205M3.3 |
| chr6_-_8102714 | 0.20 |
ENST00000502429.1 ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_-_154335300 | 0.19 |
ENST00000325926.3 |
RPRM |
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr2_+_90192768 | 0.19 |
ENST00000390275.2 |
IGKV1D-13 |
immunoglobulin kappa variable 1D-13 |
| chr11_+_86748863 | 0.19 |
ENST00000340353.7 |
TMEM135 |
transmembrane protein 135 |
| chr19_-_3500635 | 0.19 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr5_-_20575959 | 0.19 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr6_+_43737939 | 0.18 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr19_-_10420459 | 0.18 |
ENST00000403352.1 ENST00000403903.3 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
| chr9_-_104198042 | 0.17 |
ENST00000374855.4 |
ALDOB |
aldolase B, fructose-bisphosphate |
| chr21_+_34398153 | 0.17 |
ENST00000382357.3 ENST00000430860.1 ENST00000333337.3 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
| chr1_-_220263096 | 0.17 |
ENST00000463953.1 ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1 |
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr12_-_12491608 | 0.17 |
ENST00000545735.1 |
MANSC1 |
MANSC domain containing 1 |
| chr15_-_85197501 | 0.16 |
ENST00000434634.2 |
WDR73 |
WD repeat domain 73 |
| chr8_+_128426535 | 0.16 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr3_+_89156799 | 0.16 |
ENST00000452448.2 ENST00000494014.1 |
EPHA3 |
EPH receptor A3 |
| chr17_+_48823975 | 0.16 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr4_-_87281224 | 0.15 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr4_+_56815102 | 0.15 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr1_-_155880672 | 0.15 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr2_+_101591314 | 0.15 |
ENST00000450763.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr4_+_95972822 | 0.14 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr8_+_1993173 | 0.14 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr18_-_5396271 | 0.13 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr17_-_29624343 | 0.13 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr1_+_32379174 | 0.13 |
ENST00000391369.1 |
AL136115.1 |
HCG2032337; PRO1848; Uncharacterized protein |
| chrY_-_24038660 | 0.12 |
ENST00000382677.3 |
RBMY1D |
RNA binding motif protein, Y-linked, family 1, member D |
| chr6_-_27782548 | 0.12 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr11_-_104905840 | 0.12 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr19_+_48497901 | 0.12 |
ENST00000339841.2 |
ELSPBP1 |
epididymal sperm binding protein 1 |
| chr6_+_118869452 | 0.11 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr7_+_108210012 | 0.11 |
ENST00000249356.3 |
DNAJB9 |
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr1_-_226926864 | 0.11 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr6_+_43738444 | 0.11 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr5_-_86534822 | 0.11 |
ENST00000445770.2 |
AC008394.1 |
Uncharacterized protein |
| chr13_-_46626847 | 0.10 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr9_+_105757590 | 0.10 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr4_+_71226468 | 0.10 |
ENST00000226460.4 |
SMR3A |
submaxillary gland androgen regulated protein 3A |
| chr10_+_133747955 | 0.10 |
ENST00000455566.1 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr2_+_173686303 | 0.09 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr19_+_48497962 | 0.09 |
ENST00000596043.1 ENST00000597519.1 |
ELSPBP1 |
epididymal sperm binding protein 1 |
| chr16_-_15180257 | 0.09 |
ENST00000540462.1 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 1.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 3.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 2.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.7 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.7 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 1.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.4 | 1.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 1.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 1.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 0.8 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.3 | 2.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 0.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.9 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 1.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 0.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.5 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.2 | 0.5 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.1 | 1.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.8 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.3 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.4 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.1 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.0 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.6 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 3.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 3.2 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 1.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:2000354 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 1.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.6 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.6 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.7 | GO:0051646 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) mitochondrion localization(GO:0051646) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.4 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.3 | 1.9 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 1.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 2.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 4.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 3.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.2 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.4 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.8 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |