Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HOXA13
Z-value: 0.81
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | HOXA13 |
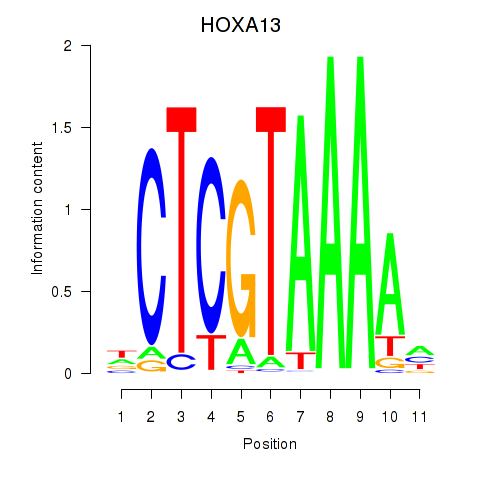
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 3.09 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr6_+_121756809 | 2.68 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr12_+_6309517 | 2.29 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr3_-_99569821 | 2.26 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr1_+_43766642 | 2.07 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr3_-_123339418 | 1.90 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr3_-_149093499 | 1.89 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr3_-_123339343 | 1.82 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr12_+_75874580 | 1.81 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr2_+_33359646 | 1.79 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chrY_+_2709527 | 1.79 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr2_+_33359687 | 1.76 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr8_-_93029865 | 1.49 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_-_91539918 | 1.49 |
ENST00000548218.1 |
DCN |
decorin |
| chr2_-_169104651 | 1.47 |
ENST00000355999.4 |
STK39 |
serine threonine kinase 39 |
| chr12_+_75874460 | 1.29 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr1_-_173176452 | 1.23 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr2_+_192109911 | 1.21 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr2_+_37571717 | 1.20 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr12_-_91546926 | 1.18 |
ENST00000550758.1 |
DCN |
decorin |
| chr22_+_38142235 | 1.15 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr14_-_92414055 | 1.08 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr9_+_706842 | 1.05 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr11_+_101983176 | 1.01 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr2_+_37571845 | 1.01 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr4_+_90823130 | 0.96 |
ENST00000508372.1 |
MMRN1 |
multimerin 1 |
| chr1_+_84609944 | 0.95 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_-_27169801 | 0.92 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr18_-_5544241 | 0.90 |
ENST00000341928.2 ENST00000540638.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr19_+_13858593 | 0.87 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr19_+_8483272 | 0.84 |
ENST00000602117.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr3_+_190333097 | 0.83 |
ENST00000412080.1 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chr17_-_46688334 | 0.81 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr6_+_148663729 | 0.81 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr9_+_91933726 | 0.76 |
ENST00000534113.2 |
SECISBP2 |
SECIS binding protein 2 |
| chr7_-_27196267 | 0.73 |
ENST00000242159.3 |
HOXA7 |
homeobox A7 |
| chr12_+_26274917 | 0.72 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr1_-_31845914 | 0.68 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr12_-_50677255 | 0.67 |
ENST00000551691.1 ENST00000394943.3 ENST00000341247.4 |
LIMA1 |
LIM domain and actin binding 1 |
| chrX_+_100878079 | 0.64 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr3_-_114477787 | 0.63 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_-_160823158 | 0.63 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr16_-_66764119 | 0.62 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr15_-_99789736 | 0.60 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr7_-_27224795 | 0.60 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr5_-_146781153 | 0.57 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr1_+_78511586 | 0.56 |
ENST00000370759.3 |
GIPC2 |
GIPC PDZ domain containing family, member 2 |
| chr4_+_74269956 | 0.56 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr16_+_82090028 | 0.55 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_-_47473425 | 0.55 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr3_-_160823040 | 0.53 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr3_-_18480260 | 0.52 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr16_+_22501658 | 0.50 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr3_+_132316081 | 0.46 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr4_-_110723134 | 0.44 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr7_-_151511911 | 0.44 |
ENST00000392801.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_+_50234309 | 0.43 |
ENST00000298307.5 |
KLHDC2 |
kelch domain containing 2 |
| chr3_-_172241250 | 0.43 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr3_-_160822858 | 0.42 |
ENST00000488170.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chrX_-_41449204 | 0.41 |
ENST00000378179.3 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr3_+_129159039 | 0.39 |
ENST00000507564.1 ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr14_-_102706934 | 0.38 |
ENST00000523231.1 ENST00000524370.1 ENST00000517966.1 |
MOK |
MOK protein kinase |
| chr3_-_8811288 | 0.38 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chr2_-_86850949 | 0.37 |
ENST00000237455.4 |
RNF103 |
ring finger protein 103 |
| chr3_-_126373929 | 0.37 |
ENST00000523403.1 ENST00000524230.2 |
TXNRD3 |
thioredoxin reductase 3 |
| chr18_+_39766626 | 0.37 |
ENST00000593234.1 ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907 |
long intergenic non-protein coding RNA 907 |
| chrX_-_38186775 | 0.36 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr10_+_114710211 | 0.35 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_-_41742697 | 0.34 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr3_-_125775629 | 0.33 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr18_-_52989217 | 0.32 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr10_+_111765562 | 0.31 |
ENST00000360162.3 |
ADD3 |
adducin 3 (gamma) |
| chr19_+_36426452 | 0.31 |
ENST00000588831.1 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
| chr7_+_80275621 | 0.31 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr22_+_18632666 | 0.31 |
ENST00000215794.7 |
USP18 |
ubiquitin specific peptidase 18 |
| chr10_-_51371321 | 0.30 |
ENST00000602930.1 |
AGAP8 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr7_+_80275752 | 0.29 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr4_-_155533787 | 0.29 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr14_-_24701539 | 0.29 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr3_-_18466026 | 0.29 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr11_-_124670550 | 0.27 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr18_-_52989525 | 0.27 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr22_+_39916558 | 0.27 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr17_-_17480779 | 0.26 |
ENST00000395782.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr2_+_162087577 | 0.26 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_+_182756615 | 0.26 |
ENST00000431877.2 ENST00000320370.7 |
SSFA2 |
sperm specific antigen 2 |
| chr9_+_137979506 | 0.26 |
ENST00000539529.1 ENST00000392991.4 ENST00000371793.3 |
OLFM1 |
olfactomedin 1 |
| chr2_+_202047596 | 0.26 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr14_-_94789663 | 0.25 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr3_+_100428188 | 0.24 |
ENST00000418917.2 ENST00000490574.1 |
TFG |
TRK-fused gene |
| chr7_+_80275953 | 0.24 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr10_+_114710425 | 0.24 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_8693539 | 0.24 |
ENST00000299663.3 |
CLEC4E |
C-type lectin domain family 4, member E |
| chr14_+_96722539 | 0.24 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr18_-_53255766 | 0.23 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr14_+_24701819 | 0.22 |
ENST00000560139.1 ENST00000559910.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr17_+_16284399 | 0.22 |
ENST00000535788.1 |
UBB |
ubiquitin B |
| chr18_+_3262954 | 0.22 |
ENST00000584539.1 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr14_+_24702073 | 0.21 |
ENST00000399440.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_+_24702127 | 0.21 |
ENST00000557854.1 ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr14_-_50319482 | 0.20 |
ENST00000546046.1 ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF |
nuclear export mediator factor |
| chr5_+_66254698 | 0.20 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_32390801 | 0.19 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr14_+_24702099 | 0.19 |
ENST00000420554.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr17_+_16284104 | 0.19 |
ENST00000577958.1 ENST00000302182.3 ENST00000577640.1 |
UBB |
ubiquitin B |
| chr4_-_89978299 | 0.18 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr3_+_101498269 | 0.17 |
ENST00000491511.2 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr4_+_70796784 | 0.17 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr16_-_18812746 | 0.16 |
ENST00000546206.2 ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1 RP11-1035H13.3 |
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr18_+_3262415 | 0.15 |
ENST00000581193.1 ENST00000400175.5 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr19_+_852291 | 0.15 |
ENST00000263621.1 |
ELANE |
elastase, neutrophil expressed |
| chr2_+_170366203 | 0.15 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chrX_+_35816459 | 0.15 |
ENST00000399988.1 ENST00000399992.1 ENST00000399987.1 ENST00000399989.1 |
MAGEB16 |
melanoma antigen family B, 16 |
| chr4_-_17513702 | 0.15 |
ENST00000428702.2 ENST00000508623.1 ENST00000513615.1 |
QDPR |
quinoid dihydropteridine reductase |
| chr19_-_44809121 | 0.14 |
ENST00000591609.1 ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235 |
zinc finger protein 235 |
| chr9_-_21305312 | 0.14 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr1_-_217804377 | 0.14 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr14_+_24701628 | 0.13 |
ENST00000355299.4 ENST00000559836.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr19_-_1592828 | 0.13 |
ENST00000592012.1 |
MBD3 |
methyl-CpG binding domain protein 3 |
| chr6_+_131894284 | 0.13 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr14_-_50319758 | 0.13 |
ENST00000298310.5 |
NEMF |
nuclear export mediator factor |
| chr16_+_66914264 | 0.13 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chrX_+_47082408 | 0.12 |
ENST00000518022.1 ENST00000276052.6 |
CDK16 |
cyclin-dependent kinase 16 |
| chr8_-_16050214 | 0.12 |
ENST00000262101.5 |
MSR1 |
macrophage scavenger receptor 1 |
| chr13_-_24895566 | 0.11 |
ENST00000422229.2 |
AL359736.1 |
protein PCOTH isoform 1 |
| chr16_-_90096309 | 0.11 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr4_+_100495864 | 0.11 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr5_+_169010638 | 0.11 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr11_+_94706804 | 0.10 |
ENST00000335080.5 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr1_-_216978709 | 0.10 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr7_-_50633078 | 0.10 |
ENST00000444124.2 |
DDC |
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr19_-_47987419 | 0.10 |
ENST00000536339.1 ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN |
kaptin (actin binding protein) |
| chr2_+_109223595 | 0.10 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr19_+_16830774 | 0.10 |
ENST00000524140.2 |
NWD1 |
NACHT and WD repeat domain containing 1 |
| chr19_-_1592652 | 0.10 |
ENST00000156825.1 ENST00000434436.3 |
MBD3 |
methyl-CpG binding domain protein 3 |
| chrX_+_37850026 | 0.10 |
ENST00000341016.3 |
CXorf27 |
chromosome X open reading frame 27 |
| chr15_+_71228826 | 0.09 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr14_+_56078695 | 0.09 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr4_+_70861647 | 0.09 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr1_+_78769549 | 0.08 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr8_-_16050288 | 0.08 |
ENST00000350896.3 |
MSR1 |
macrophage scavenger receptor 1 |
| chr19_+_16999654 | 0.08 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr6_-_167571817 | 0.08 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr12_+_120972606 | 0.08 |
ENST00000413266.2 |
RNF10 |
ring finger protein 10 |
| chr4_+_72052964 | 0.07 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_-_46769261 | 0.07 |
ENST00000343304.6 |
LRRC41 |
leucine rich repeat containing 41 |
| chr16_+_75252878 | 0.07 |
ENST00000361017.4 |
CTRB1 |
chymotrypsinogen B1 |
| chr14_-_24911868 | 0.07 |
ENST00000554698.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr17_+_16284604 | 0.07 |
ENST00000395839.1 ENST00000395837.1 |
UBB |
ubiquitin B |
| chr19_+_15838834 | 0.07 |
ENST00000305899.3 |
OR10H2 |
olfactory receptor, family 10, subfamily H, member 2 |
| chr19_-_18632861 | 0.06 |
ENST00000262809.4 |
ELL |
elongation factor RNA polymerase II |
| chr1_-_46642154 | 0.06 |
ENST00000540385.1 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr12_+_20963632 | 0.05 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr20_+_37590942 | 0.05 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr9_-_2844058 | 0.05 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr11_-_114466471 | 0.05 |
ENST00000424261.2 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr17_-_79829190 | 0.05 |
ENST00000581876.1 ENST00000584461.1 ENST00000583868.1 ENST00000400721.4 ENST00000269321.7 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr21_+_39644395 | 0.05 |
ENST00000398934.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr19_-_51340469 | 0.05 |
ENST00000326856.4 |
KLK15 |
kallikrein-related peptidase 15 |
| chr11_-_47447970 | 0.05 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr10_+_106013923 | 0.04 |
ENST00000539281.1 |
GSTO1 |
glutathione S-transferase omega 1 |
| chr15_-_55881135 | 0.04 |
ENST00000302000.6 |
PYGO1 |
pygopus family PHD finger 1 |
| chrX_-_20236970 | 0.04 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr17_-_67264947 | 0.04 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr12_+_20963647 | 0.03 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr11_+_61015594 | 0.03 |
ENST00000451616.2 |
PGA5 |
pepsinogen 5, group I (pepsinogen A) |
| chr3_-_112127981 | 0.03 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr11_+_113779704 | 0.03 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr19_+_19322758 | 0.02 |
ENST00000252575.6 |
NCAN |
neurocan |
| chrX_+_49687216 | 0.02 |
ENST00000376088.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr12_+_12878829 | 0.01 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr17_-_39538550 | 0.01 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr14_+_61201445 | 0.01 |
ENST00000261245.4 ENST00000539616.2 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
| chr8_+_133879193 | 0.01 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr4_+_70146217 | 0.01 |
ENST00000335568.5 ENST00000511240.1 |
UGT2B28 |
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr3_+_46618727 | 0.01 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr21_+_39644305 | 0.01 |
ENST00000398930.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr13_-_36429763 | 0.01 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr3_-_178969403 | 0.00 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr2_-_178937478 | 0.00 |
ENST00000286063.6 |
PDE11A |
phosphodiesterase 11A |
| chr1_-_109940550 | 0.00 |
ENST00000256637.6 |
SORT1 |
sortilin 1 |
| chr5_+_140180635 | 0.00 |
ENST00000522353.2 ENST00000532566.2 |
PCDHA3 |
protocadherin alpha 3 |
| chr11_+_120971882 | 0.00 |
ENST00000392793.1 |
TECTA |
tectorin alpha |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 2.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 1.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 3.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 3.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 4.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 2.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 2.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.7 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 2.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.5 | 1.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.3 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 1.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.3 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 2.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0046935 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 1.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 5.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.9 | 6.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 2.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.5 | 1.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.4 | 1.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 3.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 1.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 3.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.2 | 0.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 2.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.8 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.6 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.4 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.1 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.0 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0052314 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.2 | GO:1903238 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 1.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.6 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.0 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |