Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
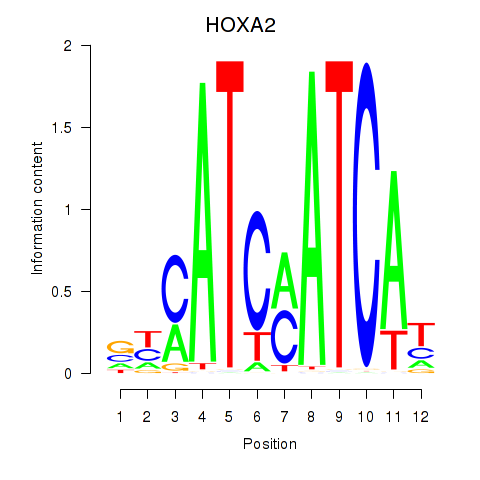
Results for HOXA2_HOXB1
Z-value: 0.96
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | HOXA2 |
|
HOXB1
|
ENSG00000120094.6 | HOXB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.42 | 1.0e-01 | Click! |
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | -0.28 | 2.9e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190044480 | 4.10 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr6_+_151561085 | 3.26 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_+_151561506 | 3.24 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chrX_-_10645773 | 2.37 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr17_-_46623441 | 2.29 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chrX_-_13835147 | 2.07 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr13_-_110438914 | 1.97 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr4_-_90757364 | 1.84 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_111782696 | 1.40 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr4_-_90756769 | 1.36 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_-_13835461 | 0.99 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr16_-_49890016 | 0.97 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr7_+_79765071 | 0.90 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr2_+_12857043 | 0.89 |
ENST00000381465.2 |
TRIB2 |
tribbles pseudokinase 2 |
| chr17_+_48609903 | 0.89 |
ENST00000268933.3 |
EPN3 |
epsin 3 |
| chr5_-_125930929 | 0.88 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr11_-_111782484 | 0.88 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr2_+_12857015 | 0.80 |
ENST00000155926.4 |
TRIB2 |
tribbles pseudokinase 2 |
| chr4_-_102267953 | 0.76 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_-_27135591 | 0.75 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr1_-_43833628 | 0.66 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr12_-_28124903 | 0.63 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_15038779 | 0.62 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr16_+_15596123 | 0.61 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr4_-_5021164 | 0.58 |
ENST00000506508.1 ENST00000509419.1 ENST00000307746.4 |
CYTL1 |
cytokine-like 1 |
| chr2_+_68961905 | 0.57 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961934 | 0.54 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr11_-_122930121 | 0.53 |
ENST00000524552.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr17_-_42277203 | 0.53 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chrX_+_99899180 | 0.50 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr10_+_124320156 | 0.49 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr10_+_124320195 | 0.47 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr6_-_159065741 | 0.45 |
ENST00000367085.3 ENST00000367089.3 |
DYNLT1 |
dynein, light chain, Tctex-type 1 |
| chr7_-_27170352 | 0.44 |
ENST00000428284.2 ENST00000360046.5 |
HOXA4 |
homeobox A4 |
| chr10_-_79397316 | 0.42 |
ENST00000372421.5 ENST00000457953.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79397202 | 0.41 |
ENST00000372437.1 ENST00000372408.2 ENST00000372403.4 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_178062855 | 0.41 |
ENST00000448150.3 |
RASAL2 |
RAS protein activator like 2 |
| chr6_-_112575912 | 0.40 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr19_+_13051206 | 0.39 |
ENST00000586760.1 |
CALR |
calreticulin |
| chr6_-_112575687 | 0.38 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr10_-_79397479 | 0.38 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_+_23386557 | 0.37 |
ENST00000523930.1 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr11_-_122929699 | 0.37 |
ENST00000526686.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr19_-_51512804 | 0.36 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr1_-_217804377 | 0.34 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr2_+_242089833 | 0.33 |
ENST00000404405.3 ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr17_+_39975544 | 0.32 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr1_+_68150744 | 0.31 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr17_+_39975455 | 0.30 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr2_-_145278475 | 0.29 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_-_109584768 | 0.28 |
ENST00000357672.3 |
WDR47 |
WD repeat domain 47 |
| chr1_-_109584608 | 0.27 |
ENST00000400794.3 ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47 |
WD repeat domain 47 |
| chr1_-_109584716 | 0.27 |
ENST00000531337.1 ENST00000529074.1 ENST00000369965.4 |
WDR47 |
WD repeat domain 47 |
| chr6_-_112575838 | 0.26 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr17_-_37353950 | 0.22 |
ENST00000394310.3 ENST00000394303.3 ENST00000344140.5 |
CACNB1 |
calcium channel, voltage-dependent, beta 1 subunit |
| chr10_+_95848824 | 0.21 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr18_-_53070913 | 0.20 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr9_+_140135665 | 0.19 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr19_-_12997995 | 0.18 |
ENST00000264834.4 |
KLF1 |
Kruppel-like factor 1 (erythroid) |
| chr2_-_45236540 | 0.18 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr6_-_112575758 | 0.17 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr3_+_189507432 | 0.17 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr7_-_14029283 | 0.16 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr9_-_73029540 | 0.15 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr1_-_67390474 | 0.15 |
ENST00000371023.3 ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78 |
WD repeat domain 78 |
| chr2_+_68962014 | 0.15 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr8_+_23386305 | 0.13 |
ENST00000519973.1 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr8_-_38008783 | 0.13 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr12_-_28125638 | 0.13 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr1_-_217250231 | 0.13 |
ENST00000493748.1 ENST00000463665.1 |
ESRRG |
estrogen-related receptor gamma |
| chr10_+_18689637 | 0.12 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_-_44588679 | 0.12 |
ENST00000409411.1 |
PREPL |
prolyl endopeptidase-like |
| chr10_+_89264625 | 0.12 |
ENST00000371996.4 ENST00000371994.4 |
MINPP1 |
multiple inositol-polyphosphate phosphatase 1 |
| chr14_+_22985251 | 0.12 |
ENST00000390510.1 |
TRAJ27 |
T cell receptor alpha joining 27 |
| chr12_+_21679220 | 0.12 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr2_-_44588694 | 0.11 |
ENST00000409957.1 |
PREPL |
prolyl endopeptidase-like |
| chr17_+_58755184 | 0.11 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr6_+_32006042 | 0.09 |
ENST00000418967.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr7_-_14029515 | 0.08 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr5_-_10761206 | 0.08 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr12_-_57472522 | 0.08 |
ENST00000379391.3 ENST00000300128.4 |
TMEM194A |
transmembrane protein 194A |
| chr12_-_114843889 | 0.08 |
ENST00000405440.2 |
TBX5 |
T-box 5 |
| chr12_-_58135903 | 0.08 |
ENST00000257897.3 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_-_86650045 | 0.07 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_+_29426230 | 0.06 |
ENST00000442615.1 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr2_+_135011731 | 0.06 |
ENST00000281923.2 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr12_+_57482877 | 0.06 |
ENST00000342556.6 ENST00000357680.4 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr12_-_86650077 | 0.05 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_+_32006159 | 0.05 |
ENST00000478281.1 ENST00000471671.1 ENST00000435122.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chrX_+_48455866 | 0.05 |
ENST00000376729.5 ENST00000218056.5 |
WDR13 |
WD repeat domain 13 |
| chr13_-_36050819 | 0.05 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr2_-_49381646 | 0.04 |
ENST00000346173.3 ENST00000406846.2 |
FSHR |
follicle stimulating hormone receptor |
| chr17_+_37824700 | 0.04 |
ENST00000581428.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr17_-_42276574 | 0.03 |
ENST00000589805.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr8_-_143999259 | 0.03 |
ENST00000323110.2 |
CYP11B2 |
cytochrome P450, family 11, subfamily B, polypeptide 2 |
| chrX_-_65253506 | 0.03 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr19_+_47421933 | 0.03 |
ENST00000404338.3 |
ARHGAP35 |
Rho GTPase activating protein 35 |
| chr2_-_49381572 | 0.02 |
ENST00000454032.1 ENST00000304421.4 |
FSHR |
follicle stimulating hormone receptor |
| chr7_-_27142290 | 0.02 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr2_-_200323414 | 0.01 |
ENST00000443023.1 |
SATB2 |
SATB homeobox 2 |
| chrX_+_95939711 | 0.01 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr1_-_223536679 | 0.01 |
ENST00000608996.1 |
SUSD4 |
sushi domain containing 4 |
| chr2_-_200322723 | 0.00 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.8 | 2.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.6 | 3.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.6 | 3.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 6.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.7 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 2.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 0.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.6 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 2.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.9 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.2 | 0.5 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 2.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.9 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 1.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.8 | GO:0048752 | embryonic neurocranium morphogenesis(GO:0048702) semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 3.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.9 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 3.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 6.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 3.1 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 6.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 5.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 1.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |