Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
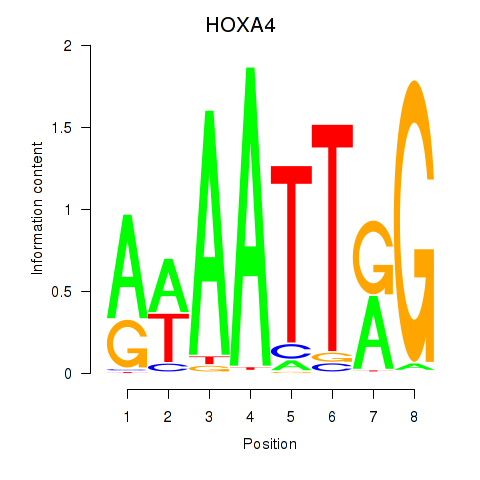
Results for HOXA4
Z-value: 0.65
Transcription factors associated with HOXA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA4
|
ENSG00000197576.9 | HOXA4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA4 | hg19_v2_chr7_-_27169801_27169844 | -0.50 | 4.8e-02 | Click! |
Activity profile of HOXA4 motif
Sorted Z-values of HOXA4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_52486841 | 1.60 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr10_-_75415825 | 1.23 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr10_-_92681033 | 1.22 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr5_-_88179302 | 1.20 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_-_190927447 | 1.09 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr17_-_10325261 | 1.02 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr5_+_152870106 | 0.92 |
ENST00000285900.5 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr16_+_6533380 | 0.86 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr13_-_46716969 | 0.83 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_-_10452929 | 0.76 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr10_+_11047259 | 0.71 |
ENST00000379261.4 ENST00000416382.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr16_+_7382745 | 0.67 |
ENST00000436368.2 ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_-_10421853 | 0.65 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr5_+_49962495 | 0.60 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_112832133 | 0.56 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr22_+_22930626 | 0.54 |
ENST00000390302.2 |
IGLV2-33 |
immunoglobulin lambda variable 2-33 (non-functional) |
| chr4_-_41884620 | 0.53 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr12_+_54447637 | 0.48 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr1_-_204329013 | 0.45 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr8_+_77593448 | 0.44 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr11_+_22688150 | 0.41 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr2_-_145278475 | 0.38 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr8_+_77593474 | 0.35 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chrX_+_37639264 | 0.35 |
ENST00000378588.4 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr2_+_54683419 | 0.33 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr8_-_95449155 | 0.30 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr15_+_80351910 | 0.30 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr3_+_88188254 | 0.30 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr17_-_39538550 | 0.28 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr1_-_42384343 | 0.26 |
ENST00000372584.1 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_+_167298281 | 0.25 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr15_+_80351977 | 0.25 |
ENST00000559157.1 ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr5_+_152870734 | 0.24 |
ENST00000521843.2 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr20_-_45035223 | 0.23 |
ENST00000450812.1 ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2 |
engulfment and cell motility 2 |
| chrX_+_129473859 | 0.23 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chrX_+_37639302 | 0.23 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chrX_-_119445306 | 0.22 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chrX_-_119445263 | 0.21 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr5_+_152870287 | 0.18 |
ENST00000340592.5 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr13_-_41593425 | 0.18 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr3_+_122103014 | 0.18 |
ENST00000232125.5 ENST00000477892.1 ENST00000469967.1 |
FAM162A |
family with sequence similarity 162, member A |
| chr6_-_26235206 | 0.17 |
ENST00000244534.5 |
HIST1H1D |
histone cluster 1, H1d |
| chr14_-_38064198 | 0.15 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr5_+_49962772 | 0.15 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_155658849 | 0.15 |
ENST00000368336.5 ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3 |
death associated protein 3 |
| chr6_+_31514622 | 0.15 |
ENST00000376146.4 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr19_+_45409011 | 0.14 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr15_+_89631381 | 0.14 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr14_-_36988882 | 0.14 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chrX_+_129473916 | 0.13 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr5_+_69321074 | 0.13 |
ENST00000380751.5 ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B |
small EDRK-rich factor 1B (centromeric) |
| chr9_+_87284675 | 0.12 |
ENST00000376208.1 ENST00000304053.6 ENST00000277120.3 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_+_89631647 | 0.12 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr2_-_163100045 | 0.12 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr12_-_57037284 | 0.12 |
ENST00000551570.1 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr17_+_37894570 | 0.12 |
ENST00000394211.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr6_-_116447283 | 0.11 |
ENST00000452729.1 ENST00000243222.4 |
COL10A1 |
collagen, type X, alpha 1 |
| chr2_-_163099885 | 0.11 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr10_+_118305435 | 0.10 |
ENST00000369221.2 |
PNLIP |
pancreatic lipase |
| chr5_+_67586465 | 0.10 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_-_56082455 | 0.10 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr13_-_28545276 | 0.10 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chr20_+_32150140 | 0.09 |
ENST00000344201.3 ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr1_+_214161272 | 0.09 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr15_+_75080883 | 0.09 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr9_+_87284622 | 0.09 |
ENST00000395882.1 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr14_-_36989336 | 0.07 |
ENST00000522719.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr14_+_39734482 | 0.07 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr3_+_69134080 | 0.07 |
ENST00000273258.3 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr7_-_140714430 | 0.07 |
ENST00000393008.3 |
MRPS33 |
mitochondrial ribosomal protein S33 |
| chr12_-_10959892 | 0.07 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr10_-_101825151 | 0.07 |
ENST00000441382.1 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr4_-_66536196 | 0.06 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr12_+_27677085 | 0.05 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chrX_-_55024967 | 0.05 |
ENST00000545676.1 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_+_40909354 | 0.05 |
ENST00000313164.9 |
C7 |
complement component 7 |
| chr6_+_50786414 | 0.05 |
ENST00000344788.3 ENST00000393655.3 ENST00000263046.4 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr17_+_73452545 | 0.05 |
ENST00000314256.7 |
KIAA0195 |
KIAA0195 |
| chr6_+_152011628 | 0.05 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr8_+_71383312 | 0.05 |
ENST00000520259.1 |
RP11-333A23.4 |
RP11-333A23.4 |
| chr14_-_80697396 | 0.04 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr8_+_22424551 | 0.04 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr12_+_53818855 | 0.04 |
ENST00000550839.1 |
AMHR2 |
anti-Mullerian hormone receptor, type II |
| chr18_+_13612613 | 0.04 |
ENST00000586765.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr18_-_28742813 | 0.03 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr17_-_10372875 | 0.03 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr11_-_62313090 | 0.03 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr3_-_188665428 | 0.03 |
ENST00000444488.1 |
TPRG1-AS1 |
TPRG1 antisense RNA 1 |
| chr11_+_12766583 | 0.03 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr1_+_245318279 | 0.02 |
ENST00000407071.2 |
KIF26B |
kinesin family member 26B |
| chr3_-_57233966 | 0.02 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr7_+_129015484 | 0.02 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr6_-_31514516 | 0.02 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr7_-_29234802 | 0.01 |
ENST00000449801.1 ENST00000409850.1 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr5_+_140535577 | 0.01 |
ENST00000539533.1 |
PCDHB17 |
Protocadherin-psi1; Uncharacterized protein |
| chr6_-_55740352 | 0.01 |
ENST00000370830.3 |
BMP5 |
bone morphogenetic protein 5 |
| chr11_+_120971882 | 0.01 |
ENST00000392793.1 |
TECTA |
tectorin alpha |
| chr5_+_159343688 | 0.00 |
ENST00000306675.3 |
ADRA1B |
adrenoceptor alpha 1B |
| chr6_-_31514333 | 0.00 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr2_+_87135076 | 0.00 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 1.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 1.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 1.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.3 | 1.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.0 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 1.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |