Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
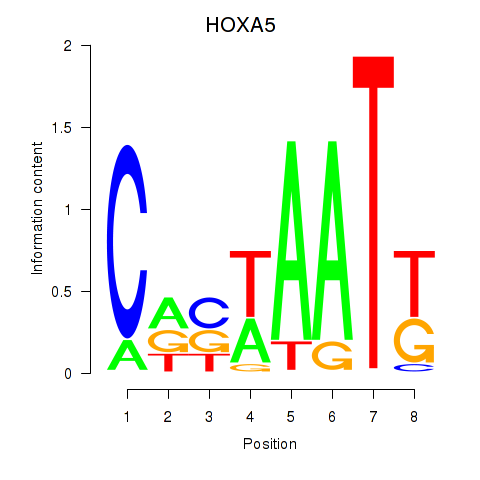
Results for HOXA5
Z-value: 0.51
Transcription factors associated with HOXA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA5
|
ENSG00000106004.4 | HOXA5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA5 | hg19_v2_chr7_-_27183263_27183287 | 0.64 | 7.4e-03 | Click! |
Activity profile of HOXA5 motif
Sorted Z-values of HOXA5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74275057 | 1.63 |
ENST00000511370.1 |
ALB |
albumin |
| chr12_-_91574142 | 1.21 |
ENST00000547937.1 |
DCN |
decorin |
| chr14_+_21156915 | 1.00 |
ENST00000397990.4 ENST00000555597.1 |
ANG RNASE4 |
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr3_-_149093499 | 0.97 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr10_-_52645379 | 0.97 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr10_-_52645416 | 0.96 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr3_-_79068594 | 0.94 |
ENST00000436010.2 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr17_+_19437132 | 0.86 |
ENST00000436810.2 ENST00000270570.4 ENST00000457293.1 ENST00000542886.1 ENST00000575023.1 ENST00000395585.1 |
SLC47A1 |
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr8_-_41166953 | 0.82 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr11_-_115375107 | 0.81 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr2_+_102508955 | 0.75 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chr11_+_12399071 | 0.71 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr1_+_100111479 | 0.69 |
ENST00000263174.4 |
PALMD |
palmdelphin |
| chr1_-_149889382 | 0.69 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr14_-_92413727 | 0.67 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr14_-_92413353 | 0.64 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr11_-_2162468 | 0.59 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_12392971 | 0.57 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr1_+_100111580 | 0.55 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chrX_+_99899180 | 0.54 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr4_+_71063641 | 0.52 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr15_-_73925651 | 0.48 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr19_+_50016610 | 0.48 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_-_116575226 | 0.45 |
ENST00000420283.1 |
TSPYL4 |
TSPY-like 4 |
| chr16_-_66764119 | 0.41 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chrX_+_54947229 | 0.41 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr1_-_227505289 | 0.37 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr11_-_46722117 | 0.34 |
ENST00000311956.4 |
ARHGAP1 |
Rho GTPase activating protein 1 |
| chr16_+_14927538 | 0.33 |
ENST00000287667.7 |
NOMO1 |
NODAL modulator 1 |
| chr16_-_18573396 | 0.30 |
ENST00000543392.1 ENST00000381474.3 ENST00000330537.6 |
NOMO2 |
NODAL modulator 2 |
| chr8_-_122653630 | 0.29 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr14_+_65878565 | 0.29 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr1_+_150337100 | 0.27 |
ENST00000401000.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_+_84609944 | 0.27 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_+_26365443 | 0.26 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr1_+_150337144 | 0.26 |
ENST00000539519.1 ENST00000369067.3 ENST00000369068.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr6_-_143266297 | 0.23 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr7_+_100136811 | 0.23 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr21_+_44073860 | 0.22 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chr19_+_18794470 | 0.22 |
ENST00000321949.8 ENST00000338797.6 |
CRTC1 |
CREB regulated transcription coactivator 1 |
| chr5_+_140723601 | 0.21 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chr11_+_9685604 | 0.21 |
ENST00000447399.2 ENST00000318950.6 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
| chr20_+_4666882 | 0.21 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr9_-_110251836 | 0.19 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr12_+_111051832 | 0.19 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr12_+_111051902 | 0.17 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr18_-_33709268 | 0.16 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr2_+_90259593 | 0.16 |
ENST00000471857.1 |
IGKV1D-8 |
immunoglobulin kappa variable 1D-8 |
| chr1_+_221051699 | 0.15 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr11_+_65686802 | 0.15 |
ENST00000376991.2 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr4_+_74606223 | 0.15 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr12_+_104337515 | 0.14 |
ENST00000550595.1 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr10_+_101089107 | 0.14 |
ENST00000446890.1 ENST00000370528.3 |
CNNM1 |
cyclin M1 |
| chr17_+_74733744 | 0.14 |
ENST00000586689.1 ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11 |
major facilitator superfamily domain containing 11 |
| chr14_+_93389425 | 0.14 |
ENST00000216492.5 ENST00000334654.4 |
CHGA |
chromogranin A (parathyroid secretory protein 1) |
| chr13_+_97874574 | 0.13 |
ENST00000343600.4 ENST00000345429.6 ENST00000376673.3 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr2_-_207082748 | 0.13 |
ENST00000407325.2 ENST00000411719.1 |
GPR1 |
G protein-coupled receptor 1 |
| chr7_-_99717463 | 0.12 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_87012753 | 0.12 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr20_-_34638841 | 0.12 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chrX_+_55478538 | 0.11 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr17_-_33446735 | 0.11 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr1_-_244006528 | 0.08 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr18_+_616672 | 0.08 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr19_+_8455077 | 0.07 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr5_+_140552218 | 0.07 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr14_-_20801427 | 0.07 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr9_-_123342415 | 0.06 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr6_+_167525277 | 0.06 |
ENST00000400926.2 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr8_-_90769422 | 0.06 |
ENST00000524190.1 ENST00000523859.1 |
RP11-37B2.1 |
RP11-37B2.1 |
| chr14_+_24025462 | 0.06 |
ENST00000556015.1 ENST00000554970.1 ENST00000554789.1 |
THTPA |
thiamine triphosphatase |
| chr20_+_58515417 | 0.05 |
ENST00000360816.3 |
FAM217B |
family with sequence similarity 217, member B |
| chr5_-_131879205 | 0.05 |
ENST00000231454.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr10_-_7708918 | 0.05 |
ENST00000256861.6 ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5 |
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr16_+_10971037 | 0.05 |
ENST00000324288.8 ENST00000381835.5 |
CIITA |
class II, major histocompatibility complex, transactivator |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_171283331 | 0.04 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chrX_+_10124977 | 0.04 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr18_+_616711 | 0.04 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr3_-_62358690 | 0.03 |
ENST00000475839.1 |
FEZF2 |
FEZ family zinc finger 2 |
| chr6_+_42896865 | 0.03 |
ENST00000372836.4 ENST00000394142.3 |
CNPY3 |
canopy FGF signaling regulator 3 |
| chr17_-_33446820 | 0.02 |
ENST00000592577.1 ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D |
RAD51 paralog D |
| chr11_-_33744256 | 0.02 |
ENST00000415002.2 ENST00000437761.2 ENST00000445143.2 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr21_-_35884573 | 0.02 |
ENST00000399286.2 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
| chr2_-_160919112 | 0.01 |
ENST00000283243.7 ENST00000392771.1 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
| chr6_+_168418553 | 0.01 |
ENST00000354419.2 ENST00000351261.3 |
KIF25 |
kinesin family member 25 |
| chr10_-_115904361 | 0.00 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr11_+_33061543 | 0.00 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr3_-_62359180 | 0.00 |
ENST00000283268.3 |
FEZF2 |
FEZ family zinc finger 2 |
| chr5_+_76326187 | 0.00 |
ENST00000312916.7 ENST00000506806.1 |
AGGF1 |
angiogenic factor with G patch and FHA domains 1 |
| chr8_-_141774467 | 0.00 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.2 | 1.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 2.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.3 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 0.9 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.3 | 0.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.2 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.3 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.7 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.1 | GO:1901899 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |