Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
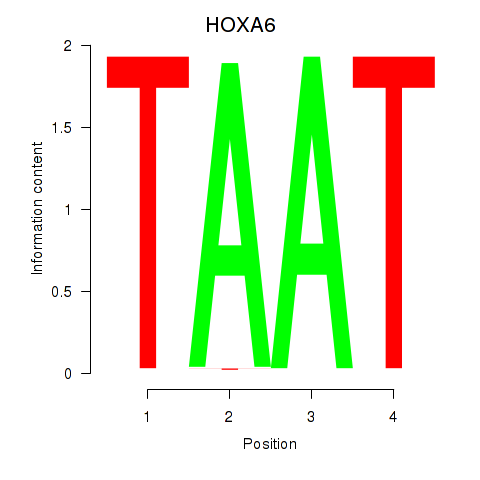
Results for HOXA6
Z-value: 0.86
Transcription factors associated with HOXA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA6
|
ENSG00000106006.6 | HOXA6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA6 | hg19_v2_chr7_-_27187393_27187393 | -0.29 | 2.8e-01 | Click! |
Activity profile of HOXA6 motif
Sorted Z-values of HOXA6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_53089723 | 4.25 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr17_-_46688334 | 2.09 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr10_+_11047259 | 2.08 |
ENST00000379261.4 ENST00000416382.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr2_-_145278475 | 2.02 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr18_-_52989217 | 1.73 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr5_-_88179302 | 1.63 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr18_-_52989525 | 1.60 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr4_+_41258786 | 1.49 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr12_-_91576750 | 1.33 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr4_+_129730779 | 1.29 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr4_+_129730947 | 1.27 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr17_-_56082455 | 1.26 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr18_-_53070913 | 1.21 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr3_+_152017181 | 1.18 |
ENST00000498502.1 ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr12_-_91576429 | 1.16 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr12_-_91576561 | 1.13 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr18_-_53177984 | 1.12 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr3_+_152017360 | 1.12 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr9_+_2159850 | 1.07 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_55095264 | 1.06 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_+_2015335 | 1.06 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_69985734 | 1.00 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr2_+_33359646 | 0.96 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.94 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr3_+_69928256 | 0.82 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr17_-_46682321 | 0.82 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr3_+_69985792 | 0.82 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr14_+_61654271 | 0.81 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr3_+_69812877 | 0.79 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_175711133 | 0.77 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr2_-_145277569 | 0.76 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_+_66662690 | 0.75 |
ENST00000488550.1 |
MEIS1 |
Meis homeobox 1 |
| chr2_+_66662510 | 0.73 |
ENST00000272369.9 ENST00000407092.2 |
MEIS1 |
Meis homeobox 1 |
| chr11_+_128562372 | 0.69 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_9431324 | 0.69 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr8_-_93107443 | 0.68 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_+_96869165 | 0.64 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_46671323 | 0.64 |
ENST00000239151.5 |
HOXB5 |
homeobox B5 |
| chr3_+_69812701 | 0.63 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr14_-_92413353 | 0.56 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr21_-_40033618 | 0.56 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr9_-_124989804 | 0.54 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr14_-_92413727 | 0.53 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr4_+_160188889 | 0.51 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_21786179 | 0.50 |
ENST00000377113.5 |
CASC10 |
cancer susceptibility candidate 10 |
| chr9_+_2158485 | 0.48 |
ENST00000417599.1 ENST00000382185.1 ENST00000382183.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_46293378 | 0.47 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr9_+_2158443 | 0.45 |
ENST00000302401.3 ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_122653630 | 0.44 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr1_+_168250194 | 0.43 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr11_+_128563652 | 0.43 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_+_141106643 | 0.41 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_88119580 | 0.41 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chrX_-_124097620 | 0.40 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr16_-_67517716 | 0.38 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr9_-_124990680 | 0.36 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr7_+_28452130 | 0.35 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr7_-_14026123 | 0.34 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr12_-_102874416 | 0.34 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr21_-_39870339 | 0.34 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr12_-_102874378 | 0.34 |
ENST00000456098.1 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr7_-_83824169 | 0.32 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_+_123396528 | 0.32 |
ENST00000322282.7 ENST00000529750.1 |
GRAMD1B |
GRAM domain containing 1B |
| chr1_+_6105974 | 0.31 |
ENST00000378083.3 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr8_-_70745575 | 0.31 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr11_-_27723158 | 0.30 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr17_+_42634844 | 0.30 |
ENST00000315323.3 |
FZD2 |
frizzled family receptor 2 |
| chr7_+_129015484 | 0.29 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr6_-_32157947 | 0.27 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr12_-_102874102 | 0.26 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr18_-_31628558 | 0.26 |
ENST00000535384.1 |
NOL4 |
nucleolar protein 4 |
| chr12_-_102591604 | 0.26 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr5_-_160973649 | 0.25 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr13_-_99667960 | 0.25 |
ENST00000448493.2 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr4_-_116034979 | 0.24 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr4_-_41884620 | 0.24 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr13_-_67802549 | 0.24 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr3_+_173302222 | 0.24 |
ENST00000361589.4 |
NLGN1 |
neuroligin 1 |
| chr11_+_20620946 | 0.23 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr8_-_95449155 | 0.22 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr1_-_28527152 | 0.22 |
ENST00000321830.5 |
AL353354.1 |
Uncharacterized protein |
| chr3_-_57233966 | 0.22 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr9_-_13165457 | 0.21 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr18_+_32073253 | 0.21 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr12_-_14133053 | 0.21 |
ENST00000609686.1 |
GRIN2B |
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr17_-_26220366 | 0.20 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr6_-_22297730 | 0.20 |
ENST00000306482.1 |
PRL |
prolactin |
| chr4_-_72649763 | 0.20 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr12_-_117799446 | 0.19 |
ENST00000317775.6 ENST00000344089.3 |
NOS1 |
nitric oxide synthase 1 (neuronal) |
| chr15_+_96876340 | 0.19 |
ENST00000453270.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_102874330 | 0.18 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr1_-_216596738 | 0.18 |
ENST00000307340.3 ENST00000366943.2 ENST00000366942.3 |
USH2A |
Usher syndrome 2A (autosomal recessive, mild) |
| chr17_-_10372875 | 0.16 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr14_+_22977587 | 0.16 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr5_+_161112754 | 0.16 |
ENST00000523217.1 |
GABRA6 |
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr7_-_14026063 | 0.15 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr15_+_34261089 | 0.15 |
ENST00000383263.5 |
CHRM5 |
cholinergic receptor, muscarinic 5 |
| chr17_-_39623681 | 0.15 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr3_+_88188254 | 0.15 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr3_+_1134260 | 0.15 |
ENST00000446702.2 ENST00000539053.1 ENST00000350110.2 |
CNTN6 |
contactin 6 |
| chrX_+_135251835 | 0.14 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr2_+_210444748 | 0.14 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr11_-_8290263 | 0.14 |
ENST00000428101.2 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr15_-_70390213 | 0.13 |
ENST00000557997.1 ENST00000317509.8 ENST00000442299.2 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr9_+_82186872 | 0.13 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_+_12538594 | 0.13 |
ENST00000543710.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_216896780 | 0.13 |
ENST00000459955.1 ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG |
estrogen-related receptor gamma |
| chr2_-_77749474 | 0.12 |
ENST00000409093.1 ENST00000409088.3 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
| chr7_-_115670804 | 0.12 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr7_-_27142290 | 0.11 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chrX_+_28605516 | 0.10 |
ENST00000378993.1 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
| chr7_-_115670792 | 0.10 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr5_-_59783882 | 0.10 |
ENST00000505507.2 ENST00000502484.2 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr15_+_58430368 | 0.10 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr9_+_2717502 | 0.10 |
ENST00000382082.3 |
KCNV2 |
potassium channel, subfamily V, member 2 |
| chr1_+_65730385 | 0.10 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr21_-_35899113 | 0.10 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr2_+_210444142 | 0.09 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr5_+_176811431 | 0.09 |
ENST00000512593.1 ENST00000324417.5 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr5_+_161112563 | 0.09 |
ENST00000274545.5 |
GABRA6 |
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr6_+_45296048 | 0.09 |
ENST00000465038.2 ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr1_+_84630053 | 0.09 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_+_115312766 | 0.09 |
ENST00000351270.3 |
HABP2 |
hyaluronan binding protein 2 |
| chr9_-_20622478 | 0.09 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr10_-_92681033 | 0.08 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr5_+_161494521 | 0.08 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chrX_+_16668278 | 0.08 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr15_+_48498480 | 0.07 |
ENST00000380993.3 ENST00000396577.3 |
SLC12A1 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_+_186265399 | 0.07 |
ENST00000367486.3 ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4 |
proteoglycan 4 |
| chr6_+_143929307 | 0.07 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr5_-_58882219 | 0.07 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr5_-_34043310 | 0.06 |
ENST00000231338.7 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
| chr13_-_103053946 | 0.06 |
ENST00000376131.4 |
FGF14 |
fibroblast growth factor 14 |
| chr7_-_25268104 | 0.06 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr22_+_39101728 | 0.05 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr15_+_80364901 | 0.05 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr4_-_176733897 | 0.05 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr18_+_32173276 | 0.05 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr6_+_42584847 | 0.05 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr5_+_161275320 | 0.05 |
ENST00000437025.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_54962100 | 0.04 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr15_+_58430567 | 0.04 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr13_-_103719196 | 0.04 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr2_+_176987088 | 0.04 |
ENST00000249499.6 |
HOXD9 |
homeobox D9 |
| chr3_-_65583561 | 0.04 |
ENST00000460329.2 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_+_161274685 | 0.04 |
ENST00000428797.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr6_+_108487245 | 0.04 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr18_+_59000815 | 0.04 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr8_-_93107827 | 0.04 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_-_93107696 | 0.03 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_+_161274940 | 0.03 |
ENST00000393943.4 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr9_-_14722715 | 0.03 |
ENST00000380911.3 |
CER1 |
cerberus 1, DAN family BMP antagonist |
| chr12_-_91398796 | 0.03 |
ENST00000261172.3 ENST00000551767.1 |
EPYC |
epiphycan |
| chr15_+_71839566 | 0.03 |
ENST00000357769.4 |
THSD4 |
thrombospondin, type I, domain containing 4 |
| chr9_-_73483958 | 0.02 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chrY_-_6742068 | 0.02 |
ENST00000215479.5 |
AMELY |
amelogenin, Y-linked |
| chr14_-_54423529 | 0.02 |
ENST00000245451.4 ENST00000559087.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr8_+_9953214 | 0.01 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr5_+_161494770 | 0.01 |
ENST00000414552.2 ENST00000361925.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_41174988 | 0.01 |
ENST00000372652.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr8_-_72268889 | 0.01 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr1_+_206557366 | 0.00 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_+_87797351 | 0.00 |
ENST00000370542.1 |
LMO4 |
LIM domain only 4 |
| chr8_+_9953061 | 0.00 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 3.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 9.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 2.0 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 1.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 3.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.9 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 1.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.2 | 1.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 2.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 4.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 1.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 10.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0002262 | myeloid cell homeostasis(GO:0002262) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 3.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.7 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.0 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.8 | 9.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 1.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 1.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 3.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 3.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 2.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0016594 | NMDA glutamate receptor activity(GO:0004972) glycine binding(GO:0016594) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |