Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HOXA9
Z-value: 0.69
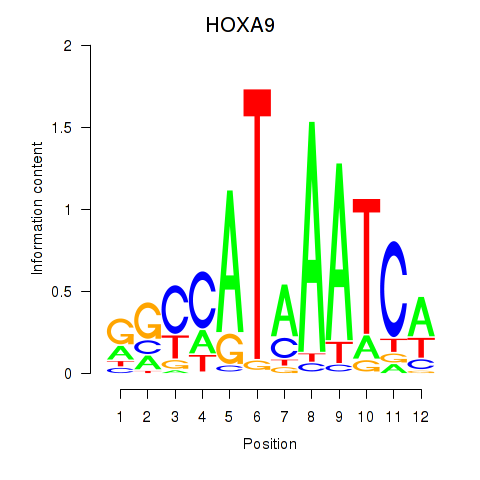
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | HOXA9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | -0.17 | 5.3e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88427568 | 2.19 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr11_-_2162162 | 1.68 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr2_+_143635067 | 1.20 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr4_-_186732048 | 1.17 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_110723194 | 1.16 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr2_+_20646824 | 1.09 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr16_-_55867146 | 1.03 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr4_-_110723134 | 1.03 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr1_-_173176452 | 0.96 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr14_-_25479811 | 0.88 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr10_-_90712520 | 0.85 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr2_+_219646462 | 0.80 |
ENST00000258415.4 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr22_+_23077065 | 0.80 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr14_-_90085458 | 0.79 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr2_+_138722028 | 0.78 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr1_+_101185290 | 0.78 |
ENST00000370119.4 ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1 |
vascular cell adhesion molecule 1 |
| chr22_+_22930626 | 0.78 |
ENST00000390302.2 |
IGLV2-33 |
immunoglobulin lambda variable 2-33 (non-functional) |
| chr13_+_98794810 | 0.77 |
ENST00000595437.1 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr4_-_70080449 | 0.73 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr2_+_143635222 | 0.72 |
ENST00000375773.2 ENST00000409512.1 ENST00000410015.2 |
KYNU |
kynureninase |
| chr18_-_10701979 | 0.71 |
ENST00000538948.1 ENST00000285141.4 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr22_+_23248512 | 0.70 |
ENST00000390325.2 |
IGLC3 |
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr3_-_8811288 | 0.69 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chr3_+_44840679 | 0.69 |
ENST00000425755.1 |
KIF15 |
kinesin family member 15 |
| chr2_+_138721850 | 0.66 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr7_+_102553430 | 0.66 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr1_-_23886285 | 0.65 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr6_+_57037089 | 0.64 |
ENST00000370693.5 |
BAG2 |
BCL2-associated athanogene 2 |
| chr3_+_157154578 | 0.60 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr15_-_72668805 | 0.57 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr1_-_89591749 | 0.53 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chr15_+_33022885 | 0.51 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr6_+_6588316 | 0.50 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr6_-_30710510 | 0.49 |
ENST00000376389.3 |
FLOT1 |
flotillin 1 |
| chrX_+_9431324 | 0.48 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr9_+_2029019 | 0.47 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_150329421 | 0.47 |
ENST00000493969.1 ENST00000328902.5 |
GIMAP6 |
GTPase, IMAP family member 6 |
| chr4_-_71532339 | 0.45 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr6_-_137539651 | 0.44 |
ENST00000543628.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr12_-_91573249 | 0.42 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr20_+_43211149 | 0.41 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_-_35441524 | 0.41 |
ENST00000395750.1 ENST00000449068.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr12_-_91573316 | 0.41 |
ENST00000393155.1 |
DCN |
decorin |
| chr16_+_12059050 | 0.40 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chrX_+_95939638 | 0.39 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr16_+_19078960 | 0.39 |
ENST00000568985.1 ENST00000566110.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr2_+_44396000 | 0.39 |
ENST00000409895.4 ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr14_+_88471468 | 0.39 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr14_-_24911448 | 0.38 |
ENST00000555355.1 ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr16_+_12058961 | 0.38 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr2_-_225811747 | 0.37 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr7_+_119913688 | 0.37 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr3_+_178276488 | 0.36 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr16_+_19079215 | 0.35 |
ENST00000544894.2 ENST00000561858.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr11_-_35440796 | 0.35 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_+_29690358 | 0.35 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr15_+_40697988 | 0.34 |
ENST00000487418.2 ENST00000479013.2 |
IVD |
isovaleryl-CoA dehydrogenase |
| chr16_-_18462221 | 0.33 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr16_-_20556492 | 0.32 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr2_+_32502952 | 0.32 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr16_+_72088376 | 0.31 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr1_+_192544857 | 0.31 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr6_+_26365443 | 0.30 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chrX_-_77150985 | 0.29 |
ENST00000358075.6 |
MAGT1 |
magnesium transporter 1 |
| chr9_-_88356789 | 0.29 |
ENST00000357081.3 ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1 |
ATP/GTP binding protein 1 |
| chr12_-_9913489 | 0.29 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr15_-_49338748 | 0.29 |
ENST00000559471.1 |
SECISBP2L |
SECIS binding protein 2-like |
| chr8_-_13372253 | 0.28 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr5_-_94417339 | 0.28 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr16_+_12059091 | 0.28 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr9_+_117373486 | 0.28 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr10_-_45474237 | 0.27 |
ENST00000448778.1 ENST00000298295.3 |
C10orf10 |
chromosome 10 open reading frame 10 |
| chr7_-_27169801 | 0.27 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr2_+_201170596 | 0.27 |
ENST00000439084.1 ENST00000409718.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr6_-_133035185 | 0.26 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr14_+_23938891 | 0.26 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr16_+_70557685 | 0.25 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr15_-_22448819 | 0.25 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr15_+_93749295 | 0.24 |
ENST00000599897.1 |
AC112693.2 |
AC112693.2 |
| chr21_+_44313375 | 0.24 |
ENST00000354250.2 ENST00000340344.4 |
NDUFV3 |
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr20_-_48782639 | 0.24 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr11_-_76155700 | 0.24 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chrX_+_95939711 | 0.24 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr12_+_25205568 | 0.23 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr14_-_106668095 | 0.23 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr2_+_208423840 | 0.23 |
ENST00000539789.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr5_-_146833222 | 0.23 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr16_-_21868739 | 0.22 |
ENST00000415645.2 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr10_-_72141330 | 0.22 |
ENST00000395011.1 ENST00000395010.1 |
LRRC20 |
leucine rich repeat containing 20 |
| chr5_-_54281491 | 0.22 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr1_+_104615595 | 0.22 |
ENST00000418362.1 |
RP11-364B6.1 |
RP11-364B6.1 |
| chr1_+_52082751 | 0.21 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr16_-_67427389 | 0.21 |
ENST00000562206.1 ENST00000290942.5 ENST00000393957.2 |
TPPP3 |
tubulin polymerization-promoting protein family member 3 |
| chr19_-_10420459 | 0.20 |
ENST00000403352.1 ENST00000403903.3 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
| chr1_-_79472365 | 0.20 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr12_+_25205666 | 0.19 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr2_+_27255806 | 0.19 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr19_-_14945933 | 0.19 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chrX_-_92928557 | 0.19 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr2_+_67624430 | 0.19 |
ENST00000272342.5 |
ETAA1 |
Ewing tumor-associated antigen 1 |
| chr16_+_19078911 | 0.18 |
ENST00000321998.5 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr1_+_101702417 | 0.18 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr17_-_56494908 | 0.18 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr3_-_9885626 | 0.17 |
ENST00000424438.1 ENST00000433555.1 ENST00000427174.1 ENST00000418713.1 ENST00000433535.2 ENST00000383820.5 ENST00000433972.1 |
RPUSD3 |
RNA pseudouridylate synthase domain containing 3 |
| chr1_-_235292250 | 0.17 |
ENST00000366607.4 |
TOMM20 |
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr18_+_11857439 | 0.17 |
ENST00000602628.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr11_-_35441597 | 0.17 |
ENST00000395753.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_234104079 | 0.17 |
ENST00000417661.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr17_-_8021710 | 0.16 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr9_-_86571628 | 0.16 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr17_-_8113542 | 0.16 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr17_+_57297807 | 0.16 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_+_148545586 | 0.16 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr11_-_76155618 | 0.16 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chrX_-_110513703 | 0.16 |
ENST00000324068.1 |
CAPN6 |
calpain 6 |
| chr20_+_57414743 | 0.16 |
ENST00000313949.7 |
GNAS |
GNAS complex locus |
| chr2_+_113239710 | 0.16 |
ENST00000233336.6 |
TTL |
tubulin tyrosine ligase |
| chr3_+_35721106 | 0.15 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_108321623 | 0.15 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr2_-_98280383 | 0.15 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr19_-_46285646 | 0.15 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr4_+_3076388 | 0.15 |
ENST00000355072.5 |
HTT |
huntingtin |
| chr1_-_36948879 | 0.15 |
ENST00000373106.1 ENST00000373104.1 ENST00000373103.1 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr8_-_59572301 | 0.15 |
ENST00000038176.3 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr19_-_50529193 | 0.15 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr18_+_32558208 | 0.15 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr14_+_58894706 | 0.14 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chr9_+_71820057 | 0.14 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr1_+_50575292 | 0.13 |
ENST00000371821.1 ENST00000371819.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr4_-_83765613 | 0.13 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chrX_+_144908928 | 0.13 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr20_+_57414795 | 0.13 |
ENST00000371098.2 ENST00000371075.3 |
GNAS |
GNAS complex locus |
| chr7_-_2272566 | 0.13 |
ENST00000402746.1 ENST00000265854.7 ENST00000429779.1 ENST00000399654.2 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr8_-_18666360 | 0.13 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr5_-_148758839 | 0.13 |
ENST00000261796.3 |
IL17B |
interleukin 17B |
| chr4_-_69215467 | 0.12 |
ENST00000579690.1 |
YTHDC1 |
YTH domain containing 1 |
| chr9_+_71819927 | 0.12 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr22_-_31364187 | 0.12 |
ENST00000215862.4 ENST00000397641.3 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr2_+_161993412 | 0.12 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_74870206 | 0.12 |
ENST00000357321.4 ENST00000349051.5 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr2_+_109204743 | 0.12 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr22_-_31324215 | 0.12 |
ENST00000429468.1 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr20_-_17539456 | 0.12 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr10_-_101190202 | 0.12 |
ENST00000543866.1 ENST00000370508.5 |
GOT1 |
glutamic-oxaloacetic transaminase 1, soluble |
| chr15_+_32774822 | 0.12 |
ENST00000593303.1 |
AC135983.2 |
Protein LOC100996413 |
| chr5_-_131347501 | 0.12 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr19_+_8455077 | 0.11 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr1_-_207095324 | 0.11 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr9_-_127269661 | 0.11 |
ENST00000373588.4 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr19_+_37341752 | 0.11 |
ENST00000586933.1 ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345 |
zinc finger protein 345 |
| chr19_-_50528584 | 0.11 |
ENST00000594092.1 ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3 |
vaccinia related kinase 3 |
| chrX_+_41583408 | 0.11 |
ENST00000302548.4 |
GPR82 |
G protein-coupled receptor 82 |
| chr5_-_131347583 | 0.11 |
ENST00000379255.1 ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_33168336 | 0.11 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr4_+_113970772 | 0.11 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr12_+_120972147 | 0.11 |
ENST00000325954.4 ENST00000542438.1 |
RNF10 |
ring finger protein 10 |
| chr12_-_3982548 | 0.10 |
ENST00000397096.2 ENST00000447133.3 ENST00000450737.2 |
PARP11 |
poly (ADP-ribose) polymerase family, member 11 |
| chr17_+_44370099 | 0.10 |
ENST00000496930.1 |
LRRC37A |
leucine rich repeat containing 37A |
| chr14_-_31926701 | 0.10 |
ENST00000310850.4 |
DTD2 |
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr6_-_25832254 | 0.10 |
ENST00000476801.1 ENST00000244527.4 ENST00000427328.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chr22_+_38142235 | 0.10 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr10_+_47894572 | 0.10 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr22_-_30642782 | 0.10 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr4_-_100356291 | 0.10 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr14_-_106453155 | 0.10 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr3_-_52713729 | 0.10 |
ENST00000296302.7 ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1 |
polybromo 1 |
| chr4_+_48018781 | 0.10 |
ENST00000295461.5 |
NIPAL1 |
NIPA-like domain containing 1 |
| chr15_+_65134088 | 0.09 |
ENST00000323544.4 ENST00000437723.1 |
PLEKHO2 AC069368.3 |
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr4_-_48018580 | 0.09 |
ENST00000514170.1 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr10_+_51572408 | 0.09 |
ENST00000374082.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chrX_+_30248553 | 0.09 |
ENST00000361644.2 |
MAGEB3 |
melanoma antigen family B, 3 |
| chr22_-_33454354 | 0.09 |
ENST00000358763.2 |
SYN3 |
synapsin III |
| chr4_-_74088800 | 0.09 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr16_+_16434185 | 0.09 |
ENST00000524823.2 |
AC138969.4 |
Protein PKD1P1 |
| chr1_-_234614849 | 0.09 |
ENST00000040877.1 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
| chr10_+_74870253 | 0.09 |
ENST00000544879.1 ENST00000537969.1 ENST00000372997.3 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr19_+_45971246 | 0.08 |
ENST00000585836.1 ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr10_+_51572339 | 0.08 |
ENST00000344348.6 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr17_+_58018269 | 0.08 |
ENST00000591035.1 |
RP11-178C3.1 |
Uncharacterized protein |
| chr6_-_74231444 | 0.08 |
ENST00000331523.2 ENST00000356303.2 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_+_118938485 | 0.08 |
ENST00000300793.6 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chrX_+_70503433 | 0.08 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr19_-_18314836 | 0.08 |
ENST00000464076.3 ENST00000222256.4 |
RAB3A |
RAB3A, member RAS oncogene family |
| chr11_-_14521379 | 0.08 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr11_-_22647350 | 0.08 |
ENST00000327470.3 |
FANCF |
Fanconi anemia, complementation group F |
| chr2_+_161993465 | 0.08 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr16_+_3074002 | 0.07 |
ENST00000326266.8 ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6 |
THO complex 6 homolog (Drosophila) |
| chr16_-_70557430 | 0.07 |
ENST00000393612.4 ENST00000564653.1 ENST00000323786.5 |
COG4 |
component of oligomeric golgi complex 4 |
| chr2_+_109204909 | 0.07 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chrX_+_65382381 | 0.07 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr10_+_124320156 | 0.07 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr11_+_71927807 | 0.07 |
ENST00000298223.6 ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2 |
folate receptor 2 (fetal) |
| chr4_-_76944621 | 0.07 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr9_-_104198042 | 0.07 |
ENST00000374855.4 |
ALDOB |
aldolase B, fructose-bisphosphate |
| chr12_-_13248705 | 0.07 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chr10_-_14050522 | 0.07 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr3_-_197686847 | 0.06 |
ENST00000265239.6 |
IQCG |
IQ motif containing G |
| chr18_-_72265035 | 0.06 |
ENST00000585279.1 ENST00000580048.1 |
LINC00909 |
long intergenic non-protein coding RNA 909 |
| chr12_-_88535747 | 0.06 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr20_-_48732472 | 0.06 |
ENST00000340309.3 ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.3 | 1.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 0.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 1.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 2.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 0.5 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.8 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.1 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.6 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.9 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 1.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 1.4 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 2.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 1.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 2.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.0 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.1 | GO:0080130 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 1.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 0.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.7 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:1990904 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |