Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
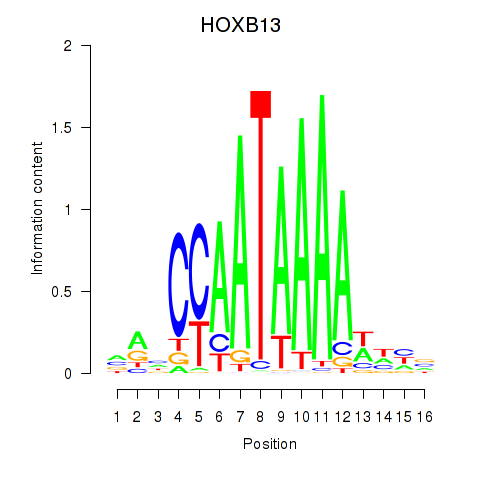
Results for HOXB13
Z-value: 0.69
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.7 | HOXB13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB13 | hg19_v2_chr17_-_46806540_46806558 | 0.18 | 5.1e-01 | Click! |
Activity profile of HOXB13 motif
Sorted Z-values of HOXB13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190044480 | 1.62 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr18_-_52989525 | 1.52 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr18_-_52989217 | 1.48 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr17_-_10560619 | 1.15 |
ENST00000583535.1 |
MYH3 |
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr2_-_175629135 | 1.06 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr3_-_114790179 | 1.00 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_+_84630645 | 0.84 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_66458072 | 0.82 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr1_-_155880672 | 0.63 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr7_-_50860565 | 0.62 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr2_-_175629164 | 0.54 |
ENST00000409323.1 ENST00000261007.5 ENST00000348749.5 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_84630053 | 0.52 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_14180778 | 0.45 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr16_-_24942411 | 0.42 |
ENST00000571843.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr1_+_23037323 | 0.41 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr16_-_24942273 | 0.37 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr15_+_66994561 | 0.35 |
ENST00000288840.5 |
SMAD6 |
SMAD family member 6 |
| chr8_+_97597148 | 0.34 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr19_+_1440838 | 0.28 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr7_-_11871815 | 0.25 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr5_-_140053152 | 0.19 |
ENST00000542735.1 |
DND1 |
DND microRNA-mediated repression inhibitor 1 |
| chr11_-_102826434 | 0.18 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr8_+_77593448 | 0.17 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr1_+_210501589 | 0.15 |
ENST00000413764.2 ENST00000541565.1 |
HHAT |
hedgehog acyltransferase |
| chr8_+_77593474 | 0.14 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr17_-_46806540 | 0.14 |
ENST00000290295.7 |
HOXB13 |
homeobox B13 |
| chr14_-_50583271 | 0.10 |
ENST00000395860.2 ENST00000395859.2 |
VCPKMT |
valosin containing protein lysine (K) methyltransferase |
| chr11_+_118401706 | 0.09 |
ENST00000411589.2 ENST00000442938.2 ENST00000359862.4 |
TMEM25 |
transmembrane protein 25 |
| chr2_+_67624430 | 0.07 |
ENST00000272342.5 |
ETAA1 |
Ewing tumor-associated antigen 1 |
| chr11_-_102714534 | 0.07 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr6_-_27782548 | 0.06 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr1_-_45956822 | 0.05 |
ENST00000372086.3 ENST00000341771.6 |
TESK2 |
testis-specific kinase 2 |
| chr4_-_164534657 | 0.05 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr18_+_3252265 | 0.05 |
ENST00000580887.1 ENST00000536605.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_-_136288113 | 0.03 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr18_+_3252206 | 0.02 |
ENST00000578562.2 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_43021612 | 0.01 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
| chr8_+_119294456 | 0.01 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr3_-_123710199 | 0.01 |
ENST00000184183.4 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr16_+_30669720 | 0.00 |
ENST00000356166.6 |
FBRS |
fibrosin |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 3.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 1.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 1.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.6 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 3.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |