Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
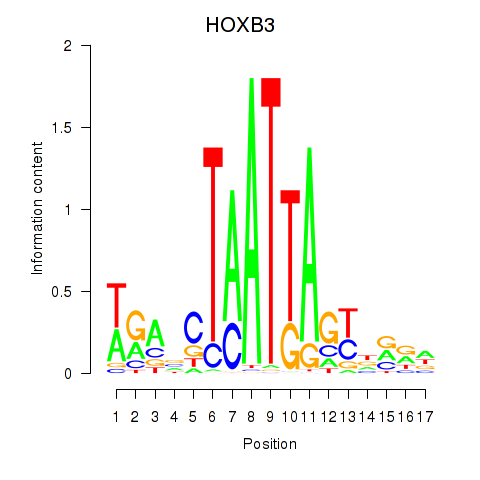
Results for HOXB3
Z-value: 1.13
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | HOXB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667594_46667619 | 0.36 | 1.7e-01 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_49834299 | 2.94 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr8_-_49833978 | 2.81 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr18_+_61554932 | 2.67 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr11_+_121447469 | 2.51 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr8_-_41166953 | 2.30 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr3_-_151034734 | 2.28 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr11_-_119991589 | 2.03 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr4_+_87515454 | 2.02 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr12_-_123201337 | 1.92 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr2_+_234637754 | 1.81 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_156030937 | 1.76 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr2_+_234545092 | 1.70 |
ENST00000344644.5 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr18_-_21891460 | 1.65 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr1_+_152486950 | 1.62 |
ENST00000368790.3 |
CRCT1 |
cysteine-rich C-terminal 1 |
| chr12_-_123187890 | 1.46 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr13_+_73632897 | 1.45 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr7_-_24797546 | 1.41 |
ENST00000414428.1 ENST00000419307.1 ENST00000342947.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chrX_+_43515467 | 1.26 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr17_-_27418537 | 1.23 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr9_+_130911723 | 1.23 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr9_+_130911770 | 1.22 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr1_-_68698222 | 1.17 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr11_-_104905840 | 1.16 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr9_+_470288 | 1.11 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr11_-_118134997 | 1.09 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr2_+_234580499 | 1.06 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234580525 | 1.04 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr18_+_55888767 | 1.03 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr7_+_142458507 | 0.98 |
ENST00000492062.1 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr5_+_36608422 | 0.92 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_-_102268484 | 0.88 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_185538849 | 0.84 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr22_+_30792980 | 0.83 |
ENST00000403484.1 ENST00000405717.3 ENST00000402592.3 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr11_-_104817919 | 0.82 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr11_-_104827425 | 0.73 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr17_+_48624450 | 0.67 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr11_-_107729887 | 0.65 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr12_-_6484376 | 0.64 |
ENST00000360168.3 ENST00000358945.3 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr11_-_102651343 | 0.62 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr1_-_68698197 | 0.61 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr5_+_150639360 | 0.58 |
ENST00000523004.1 |
GM2A |
GM2 ganglioside activator |
| chr11_+_5710919 | 0.57 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr1_+_79115503 | 0.55 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr2_+_234545148 | 0.54 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr12_+_15699286 | 0.50 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr7_+_26331541 | 0.47 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr3_-_126327398 | 0.43 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr3_+_130745688 | 0.42 |
ENST00000510769.1 ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11 |
NIMA-related kinase 11 |
| chr1_+_144173162 | 0.40 |
ENST00000356801.6 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr2_+_29336855 | 0.40 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr5_+_115177178 | 0.40 |
ENST00000316788.7 |
AP3S1 |
adaptor-related protein complex 3, sigma 1 subunit |
| chr16_-_11036300 | 0.38 |
ENST00000331808.4 |
DEXI |
Dexi homolog (mouse) |
| chr14_-_70883708 | 0.37 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr3_-_151047327 | 0.35 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr7_-_134896234 | 0.34 |
ENST00000354475.4 ENST00000344400.5 |
WDR91 |
WD repeat domain 91 |
| chr2_+_200820494 | 0.33 |
ENST00000435773.2 |
C2orf47 |
chromosome 2 open reading frame 47 |
| chr1_-_146040968 | 0.31 |
ENST00000401010.3 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr1_+_81771806 | 0.30 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr2_+_113816215 | 0.29 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
| chrX_-_13835147 | 0.28 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr2_+_113816685 | 0.27 |
ENST00000393200.2 |
IL36RN |
interleukin 36 receptor antagonist |
| chr16_+_28763108 | 0.25 |
ENST00000357796.3 ENST00000550983.1 |
NPIPB9 |
nuclear pore complex interacting protein family, member B9 |
| chr6_+_160542821 | 0.23 |
ENST00000366963.4 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_+_28261492 | 0.22 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chrX_+_135730373 | 0.22 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chrX_+_135730297 | 0.22 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr14_-_24711806 | 0.21 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_-_128894053 | 0.21 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr7_-_99277610 | 0.21 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr12_+_28410128 | 0.20 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr4_+_175204818 | 0.20 |
ENST00000503780.1 |
CEP44 |
centrosomal protein 44kDa |
| chr6_+_160542870 | 0.20 |
ENST00000324965.4 ENST00000457470.2 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr14_-_24711865 | 0.20 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_+_94706973 | 0.20 |
ENST00000536741.1 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr12_+_66582919 | 0.19 |
ENST00000545837.1 ENST00000457197.2 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
| chr1_+_207226574 | 0.19 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr11_+_3819049 | 0.19 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr3_+_121774202 | 0.19 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr6_+_149721495 | 0.18 |
ENST00000326669.4 |
SUMO4 |
small ubiquitin-like modifier 4 |
| chrX_-_24690771 | 0.18 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr20_-_44600810 | 0.18 |
ENST00000322927.2 ENST00000426788.1 |
ZNF335 |
zinc finger protein 335 |
| chr1_+_160160346 | 0.18 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_220267429 | 0.17 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr9_+_131062367 | 0.17 |
ENST00000601297.1 |
AL359091.2 |
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr1_+_158978768 | 0.17 |
ENST00000447473.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr8_-_17579726 | 0.16 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr6_+_45296391 | 0.16 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr4_-_120243545 | 0.16 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr17_+_46970134 | 0.16 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_-_70780572 | 0.16 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr1_-_93257951 | 0.16 |
ENST00000543509.1 ENST00000370331.1 ENST00000540033.1 |
EVI5 |
ecotropic viral integration site 5 |
| chr17_+_46970178 | 0.15 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr16_-_28937027 | 0.15 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr12_+_22778009 | 0.15 |
ENST00000266517.4 ENST00000335148.3 |
ETNK1 |
ethanolamine kinase 1 |
| chr6_+_26501449 | 0.14 |
ENST00000244513.6 |
BTN1A1 |
butyrophilin, subfamily 1, member A1 |
| chr7_+_129015484 | 0.14 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr8_+_27168988 | 0.14 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr7_-_134855517 | 0.14 |
ENST00000430372.1 |
C7orf49 |
chromosome 7 open reading frame 49 |
| chr14_+_69865401 | 0.14 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr1_+_160160283 | 0.14 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_+_22132810 | 0.14 |
ENST00000356766.6 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr12_+_122688090 | 0.13 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr17_+_46970127 | 0.13 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_+_138722028 | 0.13 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr9_-_5339873 | 0.13 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr16_+_28648975 | 0.13 |
ENST00000529716.1 |
NPIPB8 |
nuclear pore complex interacting protein family, member B8 |
| chr16_-_28374829 | 0.12 |
ENST00000532254.1 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
| chr21_-_27423339 | 0.12 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr11_+_94706804 | 0.12 |
ENST00000335080.5 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr14_-_24711470 | 0.12 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr14_+_22320634 | 0.12 |
ENST00000390435.1 |
TRAV8-3 |
T cell receptor alpha variable 8-3 |
| chr16_-_30257122 | 0.12 |
ENST00000520915.1 |
RP11-347C12.1 |
Putative NPIP-like protein LOC613037 |
| chrX_+_37208540 | 0.12 |
ENST00000466533.1 ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1 TM4SF2 |
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr2_+_54350316 | 0.12 |
ENST00000606865.1 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr12_+_18414446 | 0.12 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr1_+_202317815 | 0.12 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_176733897 | 0.11 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr9_+_125132803 | 0.11 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_22132847 | 0.11 |
ENST00000521356.1 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr9_+_125133315 | 0.11 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr6_-_133035185 | 0.10 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr4_-_10686475 | 0.10 |
ENST00000226951.6 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr1_-_67266939 | 0.10 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr14_-_54908043 | 0.10 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr2_-_10587897 | 0.09 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr3_+_40518599 | 0.09 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr16_-_21431078 | 0.09 |
ENST00000458643.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chrX_+_106045891 | 0.09 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr3_+_121902511 | 0.09 |
ENST00000490131.1 |
CASR |
calcium-sensing receptor |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr12_+_22778116 | 0.09 |
ENST00000538218.1 |
ETNK1 |
ethanolamine kinase 1 |
| chr3_-_44519131 | 0.09 |
ENST00000425708.2 ENST00000396077.2 |
ZNF445 |
zinc finger protein 445 |
| chr10_+_53806501 | 0.09 |
ENST00000373975.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr17_-_40337470 | 0.08 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr12_+_56521840 | 0.08 |
ENST00000394048.5 |
ESYT1 |
extended synaptotagmin-like protein 1 |
| chr16_-_21868739 | 0.08 |
ENST00000415645.2 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr6_-_42690312 | 0.08 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr5_-_22853429 | 0.08 |
ENST00000504376.2 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
| chr16_-_29415350 | 0.07 |
ENST00000524087.1 |
NPIPB11 |
nuclear pore complex interacting protein family, member B11 |
| chr3_+_148508845 | 0.07 |
ENST00000491148.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr14_+_32798547 | 0.07 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr7_+_100860949 | 0.06 |
ENST00000305105.2 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
| chr14_+_32798462 | 0.06 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chrX_+_138612889 | 0.06 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr2_+_166095898 | 0.06 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_-_8873531 | 0.06 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chrX_+_144908928 | 0.06 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr16_-_21868978 | 0.05 |
ENST00000357370.5 ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chrX_-_48824793 | 0.05 |
ENST00000376477.1 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr11_+_15095108 | 0.05 |
ENST00000324229.6 ENST00000533448.1 |
CALCB |
calcitonin-related polypeptide beta |
| chr6_+_26087509 | 0.05 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr11_-_117748138 | 0.05 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr16_-_21436459 | 0.05 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr2_+_65215604 | 0.05 |
ENST00000531327.1 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr6_+_29429217 | 0.04 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr12_-_117175819 | 0.04 |
ENST00000261318.3 ENST00000536380.1 |
C12orf49 |
chromosome 12 open reading frame 49 |
| chr1_-_51796987 | 0.04 |
ENST00000262676.5 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr7_+_5919458 | 0.04 |
ENST00000416608.1 |
OCM |
oncomodulin |
| chr15_+_64680003 | 0.04 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr11_+_33061543 | 0.03 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr19_+_48949030 | 0.03 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr2_-_120282070 | 0.03 |
ENST00000019103.5 |
SCTR |
secretin receptor |
| chr6_-_62996066 | 0.03 |
ENST00000281156.4 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
| chr19_-_3985455 | 0.03 |
ENST00000309311.6 |
EEF2 |
eukaryotic translation elongation factor 2 |
| chr21_+_43823983 | 0.03 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr12_-_91451758 | 0.03 |
ENST00000266719.3 |
KERA |
keratocan |
| chr17_-_73258425 | 0.02 |
ENST00000578348.1 ENST00000582486.1 ENST00000582717.1 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr19_-_46088068 | 0.02 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr9_-_95166841 | 0.02 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr14_-_23058063 | 0.02 |
ENST00000538631.1 ENST00000543337.1 ENST00000250498.4 |
DAD1 |
defender against cell death 1 |
| chr12_-_118796910 | 0.02 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr4_-_66536196 | 0.02 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr5_-_35089722 | 0.02 |
ENST00000511486.1 ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR |
prolactin receptor |
| chr19_+_49990811 | 0.02 |
ENST00000391857.4 ENST00000467825.2 |
RPL13A |
ribosomal protein L13a |
| chr22_-_30234218 | 0.02 |
ENST00000307790.3 ENST00000542393.1 ENST00000397771.2 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
| chr8_-_141810634 | 0.01 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chrX_-_130423200 | 0.01 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr12_+_64798095 | 0.01 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr2_-_89160770 | 0.01 |
ENST00000390240.2 |
IGKJ3 |
immunoglobulin kappa joining 3 |
| chr21_-_37914898 | 0.01 |
ENST00000399136.1 |
CLDN14 |
claudin 14 |
| chr2_-_73869508 | 0.01 |
ENST00000272425.3 |
NAT8 |
N-acetyltransferase 8 (GCN5-related, putative) |
| chr12_-_10959892 | 0.01 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr19_+_12848299 | 0.01 |
ENST00000357332.3 |
ASNA1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr2_-_20251744 | 0.01 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr16_-_66864806 | 0.00 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr6_-_32095968 | 0.00 |
ENST00000375203.3 ENST00000375201.4 |
ATF6B |
activating transcription factor 6 beta |
| chr12_-_10978957 | 0.00 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr1_-_54356404 | 0.00 |
ENST00000539954.1 |
YIPF1 |
Yip1 domain family, member 1 |
| chr2_+_28618532 | 0.00 |
ENST00000545753.1 |
FOSL2 |
FOS-like antigen 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.8 | 2.5 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.8 | 2.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.6 | 6.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.6 | 1.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 2.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 0.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 1.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 1.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 2.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.8 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.6 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 2.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.2 | GO:0042109 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0071874 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.4 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.9 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 2.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 2.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 0.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 2.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 5.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 3.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 2.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 6.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 2.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 4.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.4 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 5.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.0 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |