Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
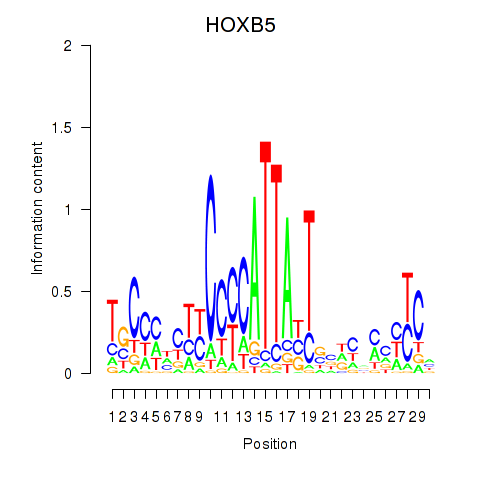
Results for HOXB5
Z-value: 0.57
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | HOXB5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg19_v2_chr17_-_46671323_46671323 | 0.43 | 9.7e-02 | Click! |
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_50143452 | 2.29 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr6_+_121756809 | 1.73 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr12_+_66217911 | 1.33 |
ENST00000403681.2 |
HMGA2 |
high mobility group AT-hook 2 |
| chr3_-_127541194 | 1.26 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chrX_+_135230712 | 1.19 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr1_+_146373546 | 1.19 |
ENST00000446760.2 |
NBPF12 |
neuroblastoma breakpoint family, member 12 |
| chr15_+_74218787 | 1.07 |
ENST00000261921.7 |
LOXL1 |
lysyl oxidase-like 1 |
| chr4_-_114900831 | 0.82 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr12_+_56915776 | 0.77 |
ENST00000550726.1 ENST00000542360.1 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chrX_+_100333709 | 0.72 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr12_+_56915713 | 0.72 |
ENST00000262031.5 ENST00000552247.2 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr3_+_51422478 | 0.68 |
ENST00000528157.1 |
MANF |
mesencephalic astrocyte-derived neurotrophic factor |
| chr12_+_53693466 | 0.49 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr15_-_75017711 | 0.45 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr14_-_102704783 | 0.44 |
ENST00000522534.1 |
MOK |
MOK protein kinase |
| chr8_-_93107443 | 0.40 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_-_188430478 | 0.38 |
ENST00000421427.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_-_141061777 | 0.38 |
ENST00000239440.4 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr5_-_141061759 | 0.34 |
ENST00000508305.1 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr8_-_93107696 | 0.27 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_+_145575980 | 0.27 |
ENST00000393045.2 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr1_+_145576007 | 0.27 |
ENST00000369298.1 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr15_-_72523454 | 0.23 |
ENST00000565154.1 ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM |
pyruvate kinase, muscle |
| chrX_+_47077632 | 0.22 |
ENST00000457458.2 |
CDK16 |
cyclin-dependent kinase 16 |
| chr2_+_17721920 | 0.21 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr6_-_32820529 | 0.21 |
ENST00000425148.2 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_+_130613226 | 0.20 |
ENST00000509662.1 ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_+_34745442 | 0.19 |
ENST00000299505.6 ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355 |
KIAA0355 |
| chr20_-_48184638 | 0.19 |
ENST00000244043.4 |
PTGIS |
prostaglandin I2 (prostacyclin) synthase |
| chr2_-_47168906 | 0.18 |
ENST00000444761.2 ENST00000409147.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr7_+_120969045 | 0.18 |
ENST00000222462.2 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
| chr12_-_6798523 | 0.18 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chrX_-_139587225 | 0.18 |
ENST00000370536.2 |
SOX3 |
SRY (sex determining region Y)-box 3 |
| chr12_-_6798616 | 0.18 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr12_-_6798410 | 0.16 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr8_-_93107827 | 0.15 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chrM_+_12331 | 0.14 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_+_46970134 | 0.11 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_46970178 | 0.10 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr8_-_133097902 | 0.08 |
ENST00000262283.5 |
OC90 |
Otoconin-90 |
| chr17_+_46970127 | 0.07 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_2496971 | 0.07 |
ENST00000397195.5 |
PAFAH1B1 |
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr5_+_149569520 | 0.05 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr12_+_54955235 | 0.05 |
ENST00000550620.1 |
PDE1B |
phosphodiesterase 1B, calmodulin-dependent |
| chr1_+_41157421 | 0.05 |
ENST00000372654.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr10_-_103603568 | 0.03 |
ENST00000356640.2 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr16_-_30381580 | 0.03 |
ENST00000409939.3 |
TBC1D10B |
TBC1 domain family, member 10B |
| chr6_-_29396243 | 0.02 |
ENST00000377148.1 |
OR11A1 |
olfactory receptor, family 11, subfamily A, member 1 |
| chrX_-_107225600 | 0.02 |
ENST00000302917.1 |
TEX13B |
testis expressed 13B |
| chr1_+_41157361 | 0.01 |
ENST00000427410.2 ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC |
nuclear transcription factor Y, gamma |
| chr18_+_32558208 | 0.01 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr20_-_3687775 | 0.01 |
ENST00000344754.4 ENST00000202578.4 |
SIGLEC1 |
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr16_-_30102547 | 0.01 |
ENST00000279386.2 |
TBX6 |
T-box 6 |
| chr1_+_155579979 | 0.00 |
ENST00000452804.2 ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1 |
misato 1, mitochondrial distribution and morphology regulator |
| chr12_-_53614155 | 0.00 |
ENST00000543726.1 |
RARG |
retinoic acid receptor, gamma |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 1.3 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.2 | 1.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.1 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 2.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 1.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 2.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.3 | 1.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |