Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
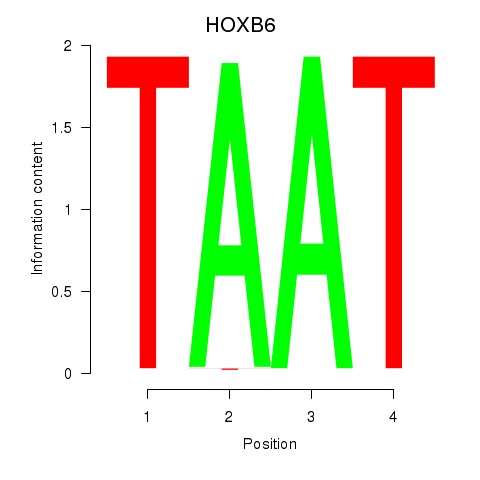
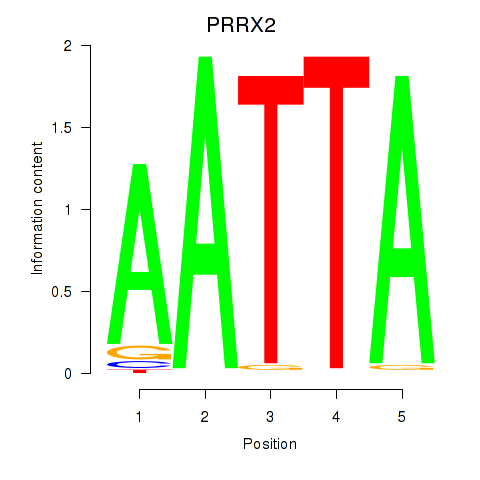
Results for HOXB6_PRRX2
Z-value: 1.92
Transcription factors associated with HOXB6_PRRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB6
|
ENSG00000108511.8 | HOXB6 |
|
PRRX2
|
ENSG00000167157.9 | PRRX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB6 | hg19_v2_chr17_-_46682321_46682362 | 0.77 | 5.1e-04 | Click! |
| PRRX2 | hg19_v2_chr9_+_132427883_132427951 | 0.19 | 4.9e-01 | Click! |
Activity profile of HOXB6_PRRX2 motif
Sorted Z-values of HOXB6_PRRX2 motif
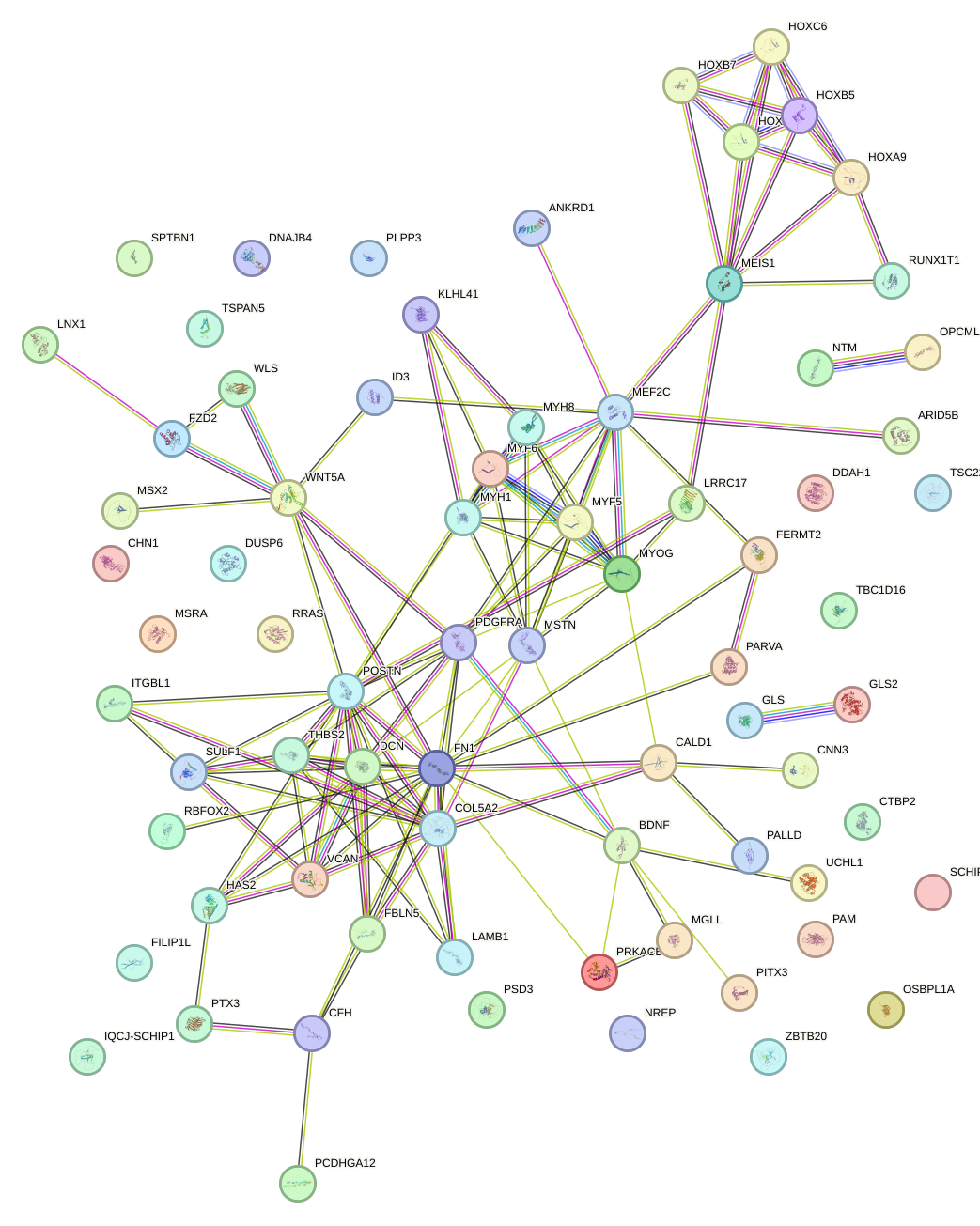
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB6_PRRX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175711133 | 9.34 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_-_27205136 | 9.07 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr12_-_91576561 | 7.89 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr12_-_91576750 | 7.75 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr18_-_53089723 | 7.25 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chrX_+_135251783 | 6.95 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr12_-_91576429 | 6.84 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr3_+_159557637 | 6.70 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr3_-_114477787 | 6.22 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_+_158787041 | 6.12 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chrX_+_135279179 | 5.82 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr7_+_102553430 | 5.57 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr3_-_114477962 | 5.54 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chrX_+_135278908 | 5.05 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chrX_+_135252050 | 4.96 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr1_-_68698222 | 4.92 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chrX_+_135251835 | 4.86 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr8_-_122653630 | 4.76 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr12_+_81110684 | 4.22 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr4_+_41258786 | 4.05 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr8_-_18744528 | 4.05 |
ENST00000523619.1 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr4_-_111563076 | 3.95 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr13_+_76334795 | 3.79 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr11_-_27723158 | 3.77 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr13_-_38172863 | 3.69 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr4_+_169418195 | 3.68 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr13_+_76334567 | 3.63 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr4_-_143226979 | 3.62 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr18_-_52989217 | 3.59 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr17_-_77924627 | 3.57 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr10_+_11047259 | 3.57 |
ENST00000379261.4 ENST00000416382.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr9_-_13165457 | 3.40 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr16_+_7382745 | 3.32 |
ENST00000436368.2 ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_-_52989525 | 3.23 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr11_+_12399071 | 3.18 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr2_-_190927447 | 3.16 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr7_+_134464376 | 3.10 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr3_-_55521323 | 3.09 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr2_-_145278475 | 3.06 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr17_-_46671323 | 3.05 |
ENST00000239151.5 |
HOXB5 |
homeobox B5 |
| chr17_-_10452929 | 3.02 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr1_-_57045228 | 2.97 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr17_-_46682321 | 2.94 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr12_-_91539918 | 2.93 |
ENST00000548218.1 |
DCN |
decorin |
| chr12_-_91574142 | 2.92 |
ENST00000547937.1 |
DCN |
decorin |
| chr13_+_102104980 | 2.89 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_91546926 | 2.83 |
ENST00000550758.1 |
DCN |
decorin |
| chr7_-_27183263 | 2.81 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr1_+_84630053 | 2.78 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_-_21891460 | 2.76 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr13_+_102104952 | 2.74 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr17_+_42634844 | 2.72 |
ENST00000315323.3 |
FZD2 |
frizzled family receptor 2 |
| chr2_+_170366203 | 2.71 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr17_-_46688334 | 2.67 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr11_+_131240373 | 2.67 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr22_-_36236623 | 2.65 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_-_92681033 | 2.62 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr5_-_111092930 | 2.62 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr2_+_54785485 | 2.61 |
ENST00000333896.5 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr2_-_216257849 | 2.59 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr17_-_10421853 | 2.58 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr14_-_92413353 | 2.57 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr7_-_107642348 | 2.54 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr3_+_157154578 | 2.53 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr2_-_151344172 | 2.50 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr7_+_134576151 | 2.49 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr7_+_134464414 | 2.48 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr6_-_169654139 | 2.47 |
ENST00000366787.3 |
THBS2 |
thrombospondin 2 |
| chr5_+_174151536 | 2.39 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr11_+_101983176 | 2.33 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr22_-_36236265 | 2.30 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr11_+_12766583 | 2.27 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr3_-_127541194 | 2.24 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr19_-_50143452 | 2.24 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr4_-_99578789 | 2.23 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr12_-_89746173 | 2.22 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr8_-_18541603 | 2.22 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr2_+_66662690 | 2.18 |
ENST00000488550.1 |
MEIS1 |
Meis homeobox 1 |
| chr12_+_81101277 | 2.15 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr14_-_92413727 | 2.12 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr13_-_45010939 | 2.11 |
ENST00000261489.2 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr1_+_84630645 | 2.11 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr13_+_102142296 | 2.10 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_-_111093167 | 2.10 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr10_+_17270214 | 2.09 |
ENST00000544301.1 |
VIM |
vimentin |
| chr17_-_10325261 | 2.09 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr5_-_111093759 | 2.07 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr1_-_95391315 | 2.02 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr2_+_66662510 | 2.00 |
ENST00000272369.9 ENST00000407092.2 |
MEIS1 |
Meis homeobox 1 |
| chr14_-_53331239 | 1.99 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr4_-_143227088 | 1.99 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr2_+_102508955 | 1.99 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chr4_+_55095264 | 1.99 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_99578776 | 1.93 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr5_+_82767487 | 1.93 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr2_-_145277569 | 1.90 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr18_-_53177984 | 1.90 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr10_+_63661053 | 1.90 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr1_-_86043921 | 1.90 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr2_+_191792376 | 1.88 |
ENST00000409428.1 ENST00000409215.1 |
GLS |
glutaminase |
| chr12_+_54422142 | 1.87 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr8_+_70404996 | 1.86 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr1_+_162602244 | 1.86 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr1_+_78470530 | 1.81 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr1_-_203055129 | 1.81 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr1_-_68698197 | 1.81 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr18_-_53070913 | 1.81 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr7_+_73442422 | 1.80 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr8_-_93107443 | 1.78 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_23886285 | 1.76 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr9_+_82186872 | 1.73 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr3_-_99569821 | 1.72 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr10_-_126716459 | 1.71 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr7_+_134576317 | 1.68 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr3_-_114343039 | 1.68 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_+_9953061 | 1.66 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr3_-_114790179 | 1.62 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr7_+_73442487 | 1.62 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr7_+_90338712 | 1.61 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr18_-_53257027 | 1.60 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr5_-_88179302 | 1.60 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr8_+_70378852 | 1.59 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr5_+_140810132 | 1.57 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr4_+_169418255 | 1.56 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr12_-_16761007 | 1.55 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr4_+_41614909 | 1.53 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr5_+_82767583 | 1.53 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr2_-_190044480 | 1.48 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_-_186877806 | 1.48 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr8_+_9953214 | 1.46 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr3_+_35721106 | 1.46 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr5_+_102201509 | 1.44 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_83072923 | 1.43 |
ENST00000535040.1 |
TPBG |
trophoblast glycoprotein |
| chr1_-_94079648 | 1.43 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr3_-_149095652 | 1.42 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr3_-_123339343 | 1.41 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr12_+_79258444 | 1.41 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr6_+_83073334 | 1.40 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr9_-_14180778 | 1.40 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr3_-_114035026 | 1.40 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr18_-_53069419 | 1.37 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr3_-_79816965 | 1.37 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr8_-_49833978 | 1.37 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr9_-_14314566 | 1.37 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr7_+_73442457 | 1.36 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr12_-_16759711 | 1.36 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_55237484 | 1.36 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr9_-_13175823 | 1.36 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr1_+_81771806 | 1.36 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr9_-_14314518 | 1.33 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr15_-_56209306 | 1.31 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr9_-_117880477 | 1.31 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr1_-_193155729 | 1.29 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr10_+_11206925 | 1.28 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr9_-_14308004 | 1.27 |
ENST00000493697.1 |
NFIB |
nuclear factor I/B |
| chrX_-_13835147 | 1.26 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr6_-_46293378 | 1.25 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr6_+_114178512 | 1.25 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr13_-_36429763 | 1.25 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr6_+_12290586 | 1.25 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr7_+_90339169 | 1.24 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr5_+_102201722 | 1.23 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr9_+_82186682 | 1.22 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr5_-_111092873 | 1.21 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr8_-_49834299 | 1.19 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr3_+_105086056 | 1.19 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr2_-_208031943 | 1.19 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr2_-_145277640 | 1.17 |
ENST00000539609.3 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_-_152589670 | 1.15 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr3_+_105085734 | 1.14 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr18_-_53253112 | 1.14 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr6_-_152639479 | 1.14 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr3_+_35722487 | 1.13 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_83824169 | 1.12 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_+_79258547 | 1.12 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr12_+_96588143 | 1.11 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr14_-_92414055 | 1.11 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr11_-_111782696 | 1.09 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_119799927 | 1.09 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr4_-_186877502 | 1.09 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_+_192141611 | 1.08 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr12_-_90049878 | 1.08 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_90049828 | 1.08 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_-_111091948 | 1.08 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr2_-_169104651 | 1.07 |
ENST00000355999.4 |
STK39 |
serine threonine kinase 39 |
| chrX_-_10645773 | 1.07 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr2_-_208030647 | 1.06 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_21593972 | 1.05 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr5_-_121413974 | 1.04 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr5_+_155753745 | 1.04 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr15_+_80351910 | 1.04 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr11_-_111794446 | 1.04 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr4_+_72204755 | 1.03 |
ENST00000512686.1 ENST00000340595.3 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr10_-_49813090 | 1.03 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr2_+_33359646 | 1.03 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_201173667 | 1.03 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr17_-_26220366 | 1.02 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr2_-_183291741 | 1.02 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_102201430 | 1.01 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr18_-_25616519 | 1.01 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 32.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 33.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 5.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 11.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 8.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 18.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 6.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 23.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.6 | 4.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.3 | 3.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 5.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 1.1 | 4.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.0 | 4.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.9 | 2.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.9 | 3.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 3.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 3.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 3.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 6.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 1.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 1.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 28.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 1.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 1.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 1.9 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 2.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 2.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 1.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 9.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 4.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 6.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 10.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 4.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.2 | 8.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 6.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 2.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 12.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 5.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.1 | 2.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 8.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 19.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 2.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 2.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.1 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 8.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 1.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 5.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 8.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 4.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0016741 | transferase activity, transferring one-carbon groups(GO:0016741) |
| 0.0 | 3.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 4.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 34.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 35.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 5.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 5.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 7.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 4.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 12.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 4.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 5.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 6.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 6.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 3.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 31.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.1 | 11.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 3.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.0 | 2.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.9 | 4.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.8 | 2.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.7 | 9.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.6 | 3.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 5.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 2.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 1.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 5.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.3 | 0.8 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 4.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 4.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 6.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 3.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 3.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 2.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 20.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 61.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 2.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 3.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 7.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 13.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 2.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 4.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.4 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0035579 | specific granule membrane(GO:0035579) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 31.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.2 | 6.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.5 | 6.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.4 | 4.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.3 | 4.0 | GO:0060577 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 1.3 | 3.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.2 | 4.9 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.2 | 3.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.1 | 4.4 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.1 | 3.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.0 | 4.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 4.8 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.9 | 2.8 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.9 | 3.7 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.9 | 2.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.9 | 2.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.8 | 5.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.8 | 2.4 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.8 | 3.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.8 | 5.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 | 8.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.7 | 3.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.7 | 4.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.7 | 2.0 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.6 | 2.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.6 | 1.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.6 | 2.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.6 | 4.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.6 | 6.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 2.7 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.5 | 1.1 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 | 2.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.5 | 10.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.5 | 27.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.5 | 1.4 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 1.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.4 | 1.8 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.4 | 2.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 4.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 12.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 3.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 0.7 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.3 | 3.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 2.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.3 | 1.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.3 | 0.7 | GO:0060437 | lung growth(GO:0060437) |
| 0.3 | 2.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 6.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 15.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 1.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 2.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.3 | 0.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 0.8 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 0.8 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.5 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.2 | 3.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.2 | 2.9 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.2 | 3.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 5.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 3.1 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 3.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.8 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.8 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 2.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.5 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.2 | 1.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.5 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.1 | 2.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 1.0 | GO:0070487 | monocyte aggregation(GO:0070487) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 2.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 5.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.9 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 2.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 1.7 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 1.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.9 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.1 | GO:1900158 | regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 2.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 1.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.3 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 2.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.2 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 3.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 22.4 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.1 | 0.2 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 2.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 3.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 3.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.2 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.2 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 | 1.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 2.0 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 5.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 1.8 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.8 | GO:0070168 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.1 | 1.6 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 3.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.6 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.2 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 2.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 2.1 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.0 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.0 | 0.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 1.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 4.3 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 3.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 2.6 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 2.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:1903539 | protein localization to postsynaptic membrane(GO:1903539) |
| 0.0 | 0.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 4.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 2.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |