Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
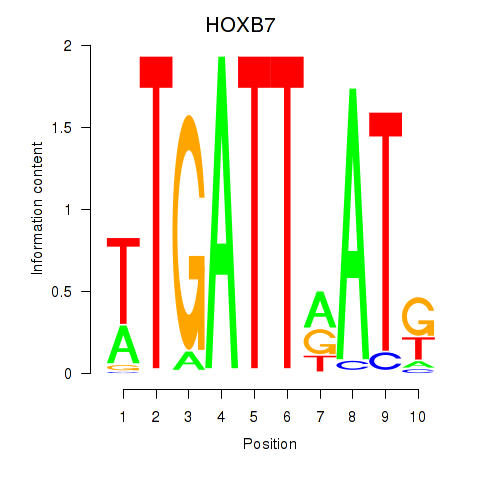
Results for HOXB7
Z-value: 0.92
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | HOXB7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | 0.10 | 7.2e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_20646824 | 2.94 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr11_-_102668879 | 2.83 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr2_-_56150910 | 2.33 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr3_+_158787041 | 2.01 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr2_-_151344172 | 1.98 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr2_-_161350305 | 1.94 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr10_-_126849068 | 1.94 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr18_+_61554932 | 1.78 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr10_+_13141585 | 1.69 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr10_+_13142225 | 1.69 |
ENST00000378747.3 |
OPTN |
optineurin |
| chrX_+_114827818 | 1.68 |
ENST00000420625.2 |
PLS3 |
plastin 3 |
| chr3_-_145878954 | 1.66 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr3_+_159557637 | 1.66 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr10_+_13142075 | 1.58 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr2_+_192141611 | 1.46 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr2_+_234637754 | 1.44 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_234621551 | 1.41 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr10_+_13141441 | 1.36 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr2_-_190044480 | 1.34 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr12_-_91546926 | 1.23 |
ENST00000550758.1 |
DCN |
decorin |
| chr9_-_32526299 | 1.10 |
ENST00000379882.1 ENST00000379883.2 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr7_-_47579188 | 1.10 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr2_+_234600253 | 1.06 |
ENST00000373424.1 ENST00000441351.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_-_54281407 | 1.02 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr12_-_28124903 | 1.00 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr1_+_152956549 | 0.99 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr4_-_102267953 | 0.96 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_+_31497271 | 0.95 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr2_+_234580525 | 0.95 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr11_+_44117260 | 0.94 |
ENST00000358681.4 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr11_+_44117099 | 0.93 |
ENST00000533608.1 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr11_+_844406 | 0.91 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr5_-_54281491 | 0.91 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr2_+_234580499 | 0.90 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234627424 | 0.90 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr14_-_74959978 | 0.82 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr5_-_125930929 | 0.82 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chrX_+_95939638 | 0.80 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chrX_-_106243451 | 0.79 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr16_+_15596123 | 0.78 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr11_-_89223883 | 0.78 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_+_101983176 | 0.78 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr2_+_102413726 | 0.78 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_-_70626430 | 0.77 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr16_+_22501658 | 0.72 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr2_+_234590556 | 0.69 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr1_-_146068184 | 0.69 |
ENST00000604894.1 ENST00000369323.3 ENST00000479926.2 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr16_-_66584059 | 0.68 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr19_+_50016610 | 0.67 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_21030260 | 0.66 |
ENST00000579303.1 |
DHRS7B |
dehydrogenase/reductase (SDR family) member 7B |
| chr14_+_55034599 | 0.66 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr12_-_91573132 | 0.60 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr13_-_110438914 | 0.58 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr1_-_147610081 | 0.58 |
ENST00000369226.3 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr8_-_18744528 | 0.58 |
ENST00000523619.1 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr1_+_19923454 | 0.58 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr2_-_188378368 | 0.57 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_-_63376013 | 0.57 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr6_+_114178512 | 0.56 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr12_-_91573249 | 0.55 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr14_+_88851874 | 0.54 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr10_+_17270214 | 0.53 |
ENST00000544301.1 |
VIM |
vimentin |
| chr16_-_66764119 | 0.52 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr9_-_129885010 | 0.52 |
ENST00000373425.3 |
ANGPTL2 |
angiopoietin-like 2 |
| chr4_-_70626314 | 0.51 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr5_+_125695805 | 0.51 |
ENST00000513040.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr10_+_71561630 | 0.51 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_-_169587621 | 0.50 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr13_-_41240717 | 0.48 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr12_-_28125638 | 0.48 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_102455846 | 0.47 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr16_-_66583701 | 0.47 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr16_-_18462221 | 0.47 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_-_102455902 | 0.47 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr11_+_2405833 | 0.46 |
ENST00000527343.1 ENST00000464784.2 |
CD81 |
CD81 molecule |
| chr5_+_140772381 | 0.46 |
ENST00000398604.2 |
PCDHGA8 |
protocadherin gamma subfamily A, 8 |
| chr12_+_56324756 | 0.44 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr11_+_57365150 | 0.44 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_-_189840223 | 0.44 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr10_+_47894572 | 0.44 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr19_-_51512804 | 0.43 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr10_+_63661053 | 0.43 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr10_-_15902449 | 0.43 |
ENST00000277632.3 |
FAM188A |
family with sequence similarity 188, member A |
| chr14_-_24584138 | 0.43 |
ENST00000558280.1 ENST00000561028.1 |
NRL |
neural retina leucine zipper |
| chr13_-_33760216 | 0.43 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr10_+_71561649 | 0.42 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr17_-_64225508 | 0.42 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr11_-_27723158 | 0.42 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr8_+_9953061 | 0.41 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr12_-_91573316 | 0.41 |
ENST00000393155.1 |
DCN |
decorin |
| chr12_-_71031185 | 0.40 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr10_+_5135981 | 0.40 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr8_+_104892639 | 0.40 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr3_+_189349162 | 0.39 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr8_+_9953214 | 0.38 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr8_+_98900132 | 0.38 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr3_-_58613323 | 0.38 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chrX_+_95939711 | 0.38 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr15_+_23255242 | 0.38 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr2_+_219110149 | 0.38 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr12_+_54674482 | 0.37 |
ENST00000547708.1 ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
| chr9_+_125132803 | 0.37 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_140227357 | 0.37 |
ENST00000378122.3 |
PCDHA9 |
protocadherin alpha 9 |
| chr19_+_52693259 | 0.37 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr14_+_62229075 | 0.36 |
ENST00000216294.4 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr14_+_74417192 | 0.36 |
ENST00000554320.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr9_+_125133315 | 0.36 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_125758865 | 0.35 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr5_+_125758813 | 0.35 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr2_+_234526272 | 0.35 |
ENST00000373450.4 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr10_+_47894023 | 0.34 |
ENST00000358474.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr15_+_84904525 | 0.33 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr15_+_85923797 | 0.32 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr4_+_148653206 | 0.32 |
ENST00000336498.3 |
ARHGAP10 |
Rho GTPase activating protein 10 |
| chr9_-_73029540 | 0.32 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr8_+_98788003 | 0.30 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr15_+_64680003 | 0.28 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr14_+_56078695 | 0.28 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr1_-_241520525 | 0.28 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr15_+_67418047 | 0.27 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr15_+_58724184 | 0.27 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr12_-_65146636 | 0.26 |
ENST00000418919.2 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
| chr4_-_110723194 | 0.26 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr19_-_52598958 | 0.24 |
ENST00000594440.1 ENST00000426391.2 ENST00000389534.4 |
ZNF841 |
zinc finger protein 841 |
| chr12_-_71031220 | 0.24 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr22_-_30642782 | 0.23 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr6_+_90272027 | 0.23 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr21_+_43619796 | 0.23 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr12_+_81471816 | 0.23 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr9_+_128509624 | 0.22 |
ENST00000342287.5 ENST00000373487.4 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr4_+_69681710 | 0.22 |
ENST00000265403.7 ENST00000458688.2 |
UGT2B10 |
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr10_+_104535994 | 0.21 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr1_+_101702417 | 0.21 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr3_+_173116225 | 0.21 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr14_-_54908043 | 0.21 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr16_-_88729473 | 0.21 |
ENST00000301012.3 ENST00000569177.1 |
MVD |
mevalonate (diphospho) decarboxylase |
| chr3_+_189507432 | 0.21 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr16_-_84220633 | 0.21 |
ENST00000566732.1 ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr12_+_56324933 | 0.20 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chrX_+_2976652 | 0.20 |
ENST00000537104.1 |
ARSF |
arylsulfatase F |
| chr11_-_13517565 | 0.20 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr7_-_36764142 | 0.20 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr8_+_39770803 | 0.20 |
ENST00000518237.1 |
IDO1 |
indoleamine 2,3-dioxygenase 1 |
| chr5_-_138534071 | 0.19 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr7_-_120498357 | 0.19 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr7_+_77325738 | 0.19 |
ENST00000334955.8 |
RSBN1L |
round spermatid basic protein 1-like |
| chr16_-_49890016 | 0.19 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr15_+_82722225 | 0.19 |
ENST00000300515.8 |
GOLGA6L9 |
golgin A6 family-like 9 |
| chr2_-_27886676 | 0.18 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr2_+_234545148 | 0.18 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr4_-_141348999 | 0.18 |
ENST00000325617.5 |
CLGN |
calmegin |
| chr17_+_43224684 | 0.18 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr8_-_29208183 | 0.18 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr12_+_12764773 | 0.18 |
ENST00000228865.2 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
| chr15_+_83098710 | 0.17 |
ENST00000561062.1 ENST00000358583.3 |
GOLGA6L9 |
golgin A6 family-like 20 |
| chr11_+_113779704 | 0.17 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr1_+_119957554 | 0.17 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr15_+_52121822 | 0.17 |
ENST00000558455.1 ENST00000308580.7 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr16_+_67880574 | 0.17 |
ENST00000219169.4 |
NUTF2 |
nuclear transport factor 2 |
| chrX_+_47053208 | 0.16 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr4_+_169013666 | 0.16 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr17_+_41363854 | 0.16 |
ENST00000588693.1 ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A |
transmembrane protein 106A |
| chr1_+_224370873 | 0.16 |
ENST00000323699.4 ENST00000391877.3 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
| chr9_+_77112244 | 0.15 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr16_-_84220604 | 0.14 |
ENST00000567759.1 |
TAF1C |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr2_-_228582709 | 0.14 |
ENST00000541617.1 ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3 |
solute carrier family 19 (thiamine transporter), member 3 |
| chrX_+_55744228 | 0.14 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr7_-_27142290 | 0.14 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr12_+_123944070 | 0.13 |
ENST00000412157.2 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr6_+_167704838 | 0.13 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr10_+_51549498 | 0.13 |
ENST00000358559.2 ENST00000298239.6 |
MSMB |
microseminoprotein, beta- |
| chr1_-_3713042 | 0.13 |
ENST00000378251.1 |
LRRC47 |
leucine rich repeat containing 47 |
| chr6_-_90024967 | 0.13 |
ENST00000602399.1 |
GABRR2 |
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr21_-_30365136 | 0.13 |
ENST00000361371.5 ENST00000389194.2 ENST00000389195.2 |
LTN1 |
listerin E3 ubiquitin protein ligase 1 |
| chr16_-_70835034 | 0.13 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chr2_+_89986318 | 0.13 |
ENST00000491977.1 |
IGKV2D-29 |
immunoglobulin kappa variable 2D-29 |
| chr2_+_113033164 | 0.13 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr9_+_140135665 | 0.12 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr13_+_21714913 | 0.12 |
ENST00000450573.1 ENST00000467636.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr8_-_110986918 | 0.12 |
ENST00000297404.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr1_-_165414414 | 0.12 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr14_+_24583836 | 0.12 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chrX_+_55744166 | 0.11 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr12_+_100897130 | 0.11 |
ENST00000551379.1 ENST00000188403.7 ENST00000551184.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr12_+_21679220 | 0.11 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr5_-_10761206 | 0.11 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chrX_+_1734051 | 0.11 |
ENST00000381229.4 ENST00000381233.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr3_+_189507460 | 0.11 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr4_-_141348789 | 0.11 |
ENST00000414773.1 |
CLGN |
calmegin |
| chr18_+_22040620 | 0.11 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr1_-_53608289 | 0.11 |
ENST00000371491.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr14_+_103851712 | 0.11 |
ENST00000440884.3 ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
| chr2_+_27255806 | 0.11 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr3_+_136581042 | 0.11 |
ENST00000288986.2 ENST00000481752.1 ENST00000491539.1 ENST00000485096.1 |
NCK1 |
NCK adaptor protein 1 |
| chr2_+_161993412 | 0.10 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_+_124103625 | 0.10 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr19_+_55315087 | 0.10 |
ENST00000345540.5 ENST00000357494.4 ENST00000396293.1 ENST00000346587.4 ENST00000396289.5 |
KIR2DL4 |
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr9_-_95166884 | 0.10 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr18_+_3252206 | 0.10 |
ENST00000578562.2 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr4_-_69536346 | 0.09 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.6 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.5 | 1.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.5 | 2.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 1.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.3 | 6.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 1.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 8.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.4 | GO:0047023 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 1.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0046625 | sphingosine-1-phosphate receptor activity(GO:0038036) sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.8 | 6.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 1.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 2.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.3 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 1.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 1.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 1.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 1.9 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 1.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.3 | 2.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 2.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 1.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.7 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 0.8 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 3.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 1.0 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.8 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.5 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.6 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.5 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.1 | 0.7 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.4 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.2 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 2.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0045918 | positive regulation of interleukin-23 production(GO:0032747) negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 1.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 1.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.9 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 4.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 6.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 2.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |