Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
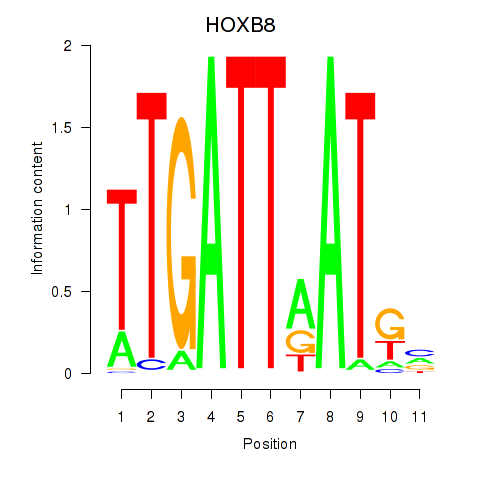
Results for HOXB8
Z-value: 1.34
Transcription factors associated with HOXB8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB8
|
ENSG00000120068.5 | HOXB8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB8 | hg19_v2_chr17_-_46690839_46690884 | -0.28 | 3.0e-01 | Click! |
Activity profile of HOXB8 motif
Sorted Z-values of HOXB8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_47173994 | 4.58 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr10_+_47746929 | 4.55 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr10_+_48255253 | 4.38 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr12_-_28124903 | 3.58 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr3_-_111314230 | 3.55 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr13_+_73629107 | 3.31 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr8_+_104892639 | 3.15 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr1_+_152956549 | 2.55 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr18_+_61554932 | 2.48 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr8_+_31497271 | 2.42 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr2_+_234580499 | 2.37 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234580525 | 2.36 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_29208183 | 2.03 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr12_-_28125638 | 2.02 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr20_-_14318248 | 2.01 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr5_+_125758865 | 1.95 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr2_+_234637754 | 1.94 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_234600253 | 1.88 |
ENST00000373424.1 ENST00000441351.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr4_-_143481822 | 1.86 |
ENST00000510812.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr11_+_5710919 | 1.84 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr3_+_158787041 | 1.84 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr19_-_10697895 | 1.81 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr18_-_21891460 | 1.76 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr2_+_234621551 | 1.67 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr5_+_125758813 | 1.66 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr4_+_169418195 | 1.65 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr10_-_126849068 | 1.62 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr2_+_192141611 | 1.61 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr8_-_49834299 | 1.56 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr20_-_56286479 | 1.55 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr3_+_189349162 | 1.53 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr12_+_56324756 | 1.46 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chrX_-_13835147 | 1.42 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr3_+_189507432 | 1.42 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr1_+_77333117 | 1.36 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr7_+_55177416 | 1.36 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr11_+_101983176 | 1.34 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr7_+_107224364 | 1.33 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr4_+_75230853 | 1.31 |
ENST00000244869.2 |
EREG |
epiregulin |
| chr13_-_110438914 | 1.29 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr4_+_169418255 | 1.24 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr10_+_13141585 | 1.21 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr12_+_113354341 | 1.20 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_+_134430212 | 1.17 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr22_+_45148432 | 1.15 |
ENST00000389774.2 ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8 |
Rho GTPase activating protein 8 |
| chr2_+_234545092 | 1.14 |
ENST00000344644.5 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_-_151344172 | 1.14 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr20_-_56265680 | 1.12 |
ENST00000414037.1 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr10_+_13141441 | 1.12 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr11_+_44117260 | 1.12 |
ENST00000358681.4 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr7_+_134528635 | 1.11 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr17_+_7942335 | 1.11 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr4_-_10041872 | 1.08 |
ENST00000309065.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr5_-_125930929 | 1.08 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr11_+_44117099 | 1.08 |
ENST00000533608.1 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr12_+_56324933 | 1.05 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr6_-_150346607 | 1.04 |
ENST00000367341.1 ENST00000286380.2 |
RAET1L |
retinoic acid early transcript 1L |
| chr1_-_6420737 | 1.01 |
ENST00000541130.1 ENST00000377845.3 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr9_-_21995300 | 0.99 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr2_+_234627424 | 0.99 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr9_+_33795533 | 0.99 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr4_-_72649763 | 0.99 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr5_+_125695805 | 0.97 |
ENST00000513040.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr2_-_161056762 | 0.96 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr5_+_36608422 | 0.91 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_+_234526272 | 0.91 |
ENST00000373450.4 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_-_161056802 | 0.90 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr3_+_30647994 | 0.89 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_+_159557637 | 0.87 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chrX_+_114827818 | 0.87 |
ENST00000420625.2 |
PLS3 |
plastin 3 |
| chr12_-_102455902 | 0.87 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr4_+_100737954 | 0.85 |
ENST00000296414.7 ENST00000512369.1 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr7_-_87856303 | 0.85 |
ENST00000394641.3 |
SRI |
sorcin |
| chr15_+_52121822 | 0.85 |
ENST00000558455.1 ENST00000308580.7 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr7_-_87856280 | 0.84 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr8_-_81083341 | 0.79 |
ENST00000519303.2 |
TPD52 |
tumor protein D52 |
| chr1_+_158985457 | 0.78 |
ENST00000567661.1 ENST00000474473.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr4_-_120243545 | 0.76 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chrX_+_102840408 | 0.75 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr1_-_237167718 | 0.75 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr11_+_62623544 | 0.73 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr2_+_219110149 | 0.73 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr3_+_87276407 | 0.72 |
ENST00000471660.1 ENST00000263780.4 ENST00000494980.1 |
CHMP2B |
charged multivesicular body protein 2B |
| chr14_-_74959994 | 0.71 |
ENST00000238633.2 ENST00000434013.2 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr12_+_20968608 | 0.70 |
ENST00000381541.3 ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3 SLCO1B3 SLCO1B7 |
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr14_-_74959978 | 0.69 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr17_+_61086917 | 0.68 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr18_+_29598335 | 0.66 |
ENST00000217740.3 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
| chr11_+_2405833 | 0.66 |
ENST00000527343.1 ENST00000464784.2 |
CD81 |
CD81 molecule |
| chr12_-_102455846 | 0.65 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr9_-_27529726 | 0.64 |
ENST00000262244.5 |
MOB3B |
MOB kinase activator 3B |
| chr5_+_140227048 | 0.64 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr9_+_130911723 | 0.63 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr21_-_43346790 | 0.62 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr9_+_130911770 | 0.62 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr10_+_13142225 | 0.61 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr11_+_62623621 | 0.60 |
ENST00000535296.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr15_-_72523454 | 0.55 |
ENST00000565154.1 ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM |
pyruvate kinase, muscle |
| chr1_+_19923454 | 0.55 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr10_+_17270214 | 0.55 |
ENST00000544301.1 |
VIM |
vimentin |
| chr19_+_58694396 | 0.53 |
ENST00000326804.4 ENST00000345813.3 ENST00000424679.2 |
ZNF274 |
zinc finger protein 274 |
| chr17_+_21030260 | 0.52 |
ENST00000579303.1 |
DHRS7B |
dehydrogenase/reductase (SDR family) member 7B |
| chr17_+_43224684 | 0.52 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr11_+_49050504 | 0.51 |
ENST00000332682.7 |
TRIM49B |
tripartite motif containing 49B |
| chr4_+_70796784 | 0.50 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr7_+_26332645 | 0.49 |
ENST00000396376.1 |
SNX10 |
sorting nexin 10 |
| chr3_-_107777208 | 0.48 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr7_-_27142290 | 0.47 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr12_+_54402790 | 0.47 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr15_-_72523924 | 0.47 |
ENST00000566809.1 ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM |
pyruvate kinase, muscle |
| chr12_+_56915776 | 0.47 |
ENST00000550726.1 ENST00000542360.1 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr16_-_10652993 | 0.46 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr2_-_161350305 | 0.46 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr19_-_14889349 | 0.46 |
ENST00000315576.3 ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr3_-_79816965 | 0.46 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr3_-_64009658 | 0.45 |
ENST00000394431.2 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr2_+_201450591 | 0.45 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr5_+_140772381 | 0.45 |
ENST00000398604.2 |
PCDHGA8 |
protocadherin gamma subfamily A, 8 |
| chr8_-_87755878 | 0.45 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr5_-_122372354 | 0.44 |
ENST00000306442.4 |
PPIC |
peptidylprolyl isomerase C (cyclophilin C) |
| chr1_-_95391315 | 0.44 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr2_+_234545148 | 0.42 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_169312350 | 0.42 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr8_+_30244580 | 0.42 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr3_-_156272924 | 0.41 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr10_+_13142075 | 0.41 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr3_-_64009102 | 0.41 |
ENST00000478185.1 ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr1_-_146068184 | 0.41 |
ENST00000604894.1 ENST00000369323.3 ENST00000479926.2 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr14_-_67826486 | 0.41 |
ENST00000555431.1 ENST00000554236.1 ENST00000555474.1 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr7_-_92747269 | 0.40 |
ENST00000446617.1 ENST00000379958.2 |
SAMD9 |
sterile alpha motif domain containing 9 |
| chr15_+_66679155 | 0.40 |
ENST00000307102.5 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
| chr1_-_147599549 | 0.40 |
ENST00000369228.5 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr2_+_102413726 | 0.40 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr9_+_128509624 | 0.40 |
ENST00000342287.5 ENST00000373487.4 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr13_-_107220455 | 0.40 |
ENST00000400198.3 |
ARGLU1 |
arginine and glutamate rich 1 |
| chr4_+_74347400 | 0.40 |
ENST00000226355.3 |
AFM |
afamin |
| chrX_-_106243451 | 0.40 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr10_-_15902449 | 0.39 |
ENST00000277632.3 |
FAM188A |
family with sequence similarity 188, member A |
| chr20_-_13619550 | 0.39 |
ENST00000455532.1 ENST00000544472.1 ENST00000539805.1 ENST00000337743.4 |
TASP1 |
taspase, threonine aspartase, 1 |
| chr15_+_41245160 | 0.38 |
ENST00000444189.2 ENST00000446533.3 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr6_+_29068386 | 0.38 |
ENST00000377171.3 |
OR2J1 |
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr8_+_98788003 | 0.38 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr8_-_116681221 | 0.38 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr13_-_30881621 | 0.37 |
ENST00000380615.3 |
KATNAL1 |
katanin p60 subunit A-like 1 |
| chr5_+_140186647 | 0.36 |
ENST00000512229.2 ENST00000356878.4 ENST00000530339.1 |
PCDHA4 |
protocadherin alpha 4 |
| chr11_+_844406 | 0.35 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr8_-_17941575 | 0.35 |
ENST00000417108.2 |
ASAH1 |
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr1_+_145292806 | 0.35 |
ENST00000448873.2 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr1_-_148347506 | 0.34 |
ENST00000369189.3 |
NBPF20 |
neuroblastoma breakpoint family, member 20 |
| chr12_-_15104040 | 0.34 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr6_+_114178512 | 0.34 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr10_+_122610687 | 0.34 |
ENST00000263461.6 |
WDR11 |
WD repeat domain 11 |
| chr3_-_55521323 | 0.34 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr5_-_178157700 | 0.34 |
ENST00000335815.2 |
ZNF354A |
zinc finger protein 354A |
| chr4_-_69536346 | 0.34 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr16_-_88729473 | 0.33 |
ENST00000301012.3 ENST00000569177.1 |
MVD |
mevalonate (diphospho) decarboxylase |
| chr4_+_88754113 | 0.33 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr19_-_14992264 | 0.33 |
ENST00000327462.2 |
OR7A17 |
olfactory receptor, family 7, subfamily A, member 17 |
| chr1_-_146082633 | 0.33 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr1_+_224544552 | 0.33 |
ENST00000465271.1 ENST00000366858.3 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
| chr16_-_66584059 | 0.33 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr3_+_105085734 | 0.33 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr11_-_122931881 | 0.33 |
ENST00000526110.1 ENST00000227378.3 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr2_-_10587897 | 0.32 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chrX_+_13707235 | 0.32 |
ENST00000464506.1 |
RAB9A |
RAB9A, member RAS oncogene family |
| chr11_+_12766583 | 0.32 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_+_103851712 | 0.32 |
ENST00000440884.3 ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
| chr1_+_224544572 | 0.32 |
ENST00000366857.5 ENST00000366856.3 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
| chr20_-_7921090 | 0.32 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr10_+_124320195 | 0.32 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr5_+_140762268 | 0.32 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr2_+_54342574 | 0.31 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr1_-_241803679 | 0.31 |
ENST00000331838.5 |
OPN3 |
opsin 3 |
| chr5_+_140213815 | 0.31 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr1_+_145301735 | 0.31 |
ENST00000605176.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr3_-_185538849 | 0.30 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr14_+_22337014 | 0.30 |
ENST00000390436.2 |
TRAV13-1 |
T cell receptor alpha variable 13-1 |
| chr11_+_117947724 | 0.30 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr8_+_9953214 | 0.30 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr5_-_9630463 | 0.30 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr2_+_207024306 | 0.30 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_64239657 | 0.30 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr8_+_9953061 | 0.29 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr19_-_52598958 | 0.29 |
ENST00000594440.1 ENST00000426391.2 ENST00000389534.4 |
ZNF841 |
zinc finger protein 841 |
| chr12_-_10962767 | 0.29 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr7_-_143454789 | 0.29 |
ENST00000470691.2 |
CTAGE6 |
CTAGE family, member 6 |
| chr9_-_70490107 | 0.29 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr16_+_15489603 | 0.29 |
ENST00000568766.1 ENST00000287594.7 |
RP11-1021N1.1 MPV17L |
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr3_+_105086056 | 0.29 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr12_+_113416191 | 0.29 |
ENST00000342315.4 ENST00000392583.2 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr11_-_128894053 | 0.28 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr7_+_143268894 | 0.28 |
ENST00000420911.2 |
CTAGE15 |
cTAGE family member 15 |
| chr14_+_61201445 | 0.27 |
ENST00000261245.4 ENST00000539616.2 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
| chr7_+_141695671 | 0.27 |
ENST00000497673.1 ENST00000475668.2 |
MGAM |
maltase-glucoamylase (alpha-glucosidase) |
| chr11_-_59633951 | 0.27 |
ENST00000257264.3 |
TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr5_+_109025067 | 0.26 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr2_+_17721230 | 0.26 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chr3_-_11645925 | 0.26 |
ENST00000413604.1 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr9_+_77112244 | 0.26 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr15_-_42565606 | 0.26 |
ENST00000307216.6 ENST00000448392.1 |
TMEM87A |
transmembrane protein 87A |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 2.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 1.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 6.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 2.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 2.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 2.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 2.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 3.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 2.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.5 | 1.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 1.9 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 13.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 4.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.1 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.3 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 3.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.6 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 5.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 5.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 2.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 2.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.6 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 3.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 4.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 3.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 1.4 | 6.9 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.0 | 1.0 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.8 | 5.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 3.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.5 | 1.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 1.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.4 | 1.3 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.4 | 3.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 3.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 3.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 2.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 0.9 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.3 | 1.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 1.3 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.3 | 1.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.0 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 1.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.3 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.2 | 2.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 2.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 5.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 1.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 1.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 0.7 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 1.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.5 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 1.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.2 | 0.5 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.5 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 2.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.7 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.3 | GO:0007605 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.0 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.4 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.3 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 1.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.4 | GO:0002836 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 2.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 1.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 2.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.5 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 1.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 2.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 1.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.3 | 7.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 5.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 6.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |